Staff Profile of Dr Georgina Brennan

- Name
- Dr Georgina Brennan
- Position
- Postdoctoral Research Officer
- g.l.brennan@bangor.ac.uk
- Phone
- 01248 382320
- Location
- School of Natural Sciences, Bangor University, Deiniol Road, Bangor, Gwynedd, LL57 2UW, UK
About
I have a strong interest in ecology and how environmental changes shape populations at the phenotypic and molecular level. I strongly believe in multidisciplinary research, combining molecular, ecological, experimental and modelling tools to best answer the question at hand.
In 2016, I joined MEEB as a postdoctoral researcher investigating the deposition of aerial pollen species with a diverse multidisciplinary group of investigators who make up team PollerGEN. We use molecular ecology, genomic approaches and ecological modelling of grass pollen in order to detect grasses to the species level, measures abundance and use this information to understand the relationship between grass pollen and asthma incidents across the UK.
My PhD, at the University of Edinburgh, explored the effect of multiple environmental changes on the effect of growth of a model organism Chlamydomonas reinhardtii. Using a selection experiment, differences between short-term and long-term responses to multiple environmental changes could be identified, as well as answering questions such as, how do populations evolve under simultaneous environmental change? And how is this different from evolving in response to a single aspect of the environment changing? Global change will involve multiple environmental changes for example in the ocean, carbon, pH, temperature, salinity, mixing etc. Understanding how microalgae, which are responsible for about half of global primary productivity, will evolve in response to multiple environmental changes is important, as their trait evolution, such as growth rate, cell size, chlorophyll content, has the potential to affect bloom events, primary production, and the species composition of aquatic communities. To read more see: http://www.smallbutmighty.bio.ed.ac.uk/.
In 2009, I completed a degree in Biology at Royal Holloway, University of London. Following my undergraduate degree, I decided to further explore molecular ecology for my Master’s degree, which I completed in 2011, at Queen’s University Belfast. For my Master’s thesis I examined the impact of the complex hydrodynamic environments on dispersal, how this structures populations of the seaweed Laminiaria digitata and the consequences to gene flow.
Follow me on Researchgate: https://www.researchgate.net/profile/Georgina_Brennan
Follow our progress on Twitter:
CV
Education
2011 – 2015 Ph.D, Institute of Evolutionary Biology, University of Edinburgh
Supervisors: Sinead Collins (PI) and Nick Colegrave (second supervisor)
Thesis: The evolutionary response of a photosynthetic green alga (Chlamydomonas reinhardtii ) to multiple environmental drivers.
2010 -2011 MSc. Molecular Ecology (Distinction), Queen’s University Belfast
Supervisor: Jim Provan
MSc Thesis: Using population genetics to understand macroalgal dispersal in a complex hydrodynamic environment.
2006 – 2009 BSc Hons. Biology (2:1), Royal Holloway, University of London
Supervisor: David Morritt
Final year research project: ‘The effect of caffeine on the common garden snail Helix aspersa.’
Conferences and workshops attended
| 2018 | Bangor University, UK. | Genomics Symposium (oral presentation) |
| 2018 | Institute of Marine Science (ICM) Barcelona, Spain | The Ramon Margalef Summer Colloquium, entitled “Ecology through the omics lens” (oral presentation). |
| 2017 | Kruger National Park, South Africa. Awarded funding by the Genetics Society. |
International Barcode of Life conference (iBOL) (oral presentation) |
| 2017 | Bangor University, UK. Invited Panellist |
Grants & Fellowships Event for Early Career Researchers |
| 2017 | Elixir, San Michele all’Adige (TN), Italy | Machine Learning for Biologists |
| 2017 | University of Birmingham, UK. |
NERC-MDIBL Environmental Genomics and Metabolomics workshop |
| 2016 | Liverpool, UK. | British Ecological Society (BES) (poster presentation) |
| 2016 | London, UK. | Linnaean Society Specialist working group in palynology (poster presentation) |
| 2016 | Edinburgh, UK. | DNA Working group meeting (oral presentation) |
2016 |
University of Leipzig, Germany |
Invited participant in workshop: synthesizing Predictability Research of Ecological Dynamics (sPRED) – funded by sDiv |
2016 |
University of Würzburg, Germany |
Metabarcoding Summer School – Scholarship awarded by DAAD |
2016 |
SCENE, Scotland |
Python for Biologists. Junior Scientist Grant – The Genetics Society |
2016 |
University of Stirling, UK |
Workshop introduction to mathematical modelling – Scholarship awarded by NERC |
2015 |
University of Lund, Sweden |
Invited speaker at the University of Lund Unit Day (Oral presentation) |
2015 |
University of Stirling, UK |
Freshwater Meeting (poster presentation) |
2015 |
University of Huddersfield, UK |
British Society for Protist Biology (Oral presentation) Student bursary awarded |
2015 |
University of Edinburgh, UK |
Edinburgh Ecology Network (EdEN) Symposium (Oral presentation) |
2014 |
New Hampshire, USA |
Ocean Global Change Biology Gordon Research Conference (Poster presentation) |
2013 |
University of Exeter, UK |
European Meeting of PhD Students in Evolutionary Biology (Oral presentation) |
2012 |
La Fouly, Switzerland |
Workshop: Bridging Theoretical and Experimental Evolution (Oral presentation) |
2012 |
University of Newcastle, UK |
British Phycological Society (Poster presentation) |
Public outreach
Publication in The Conversation: Creer, S. and Brennan, G. (2018) We’re working on a more accurate pollen forecasting system using plant DNA, The Conversation. https://theconversation.com/were-working-on-a-more-accurate-pollen-forecasting-system-using-plant-dna-97474.
Podcastfor Methods in Ecology and Evolution: The Field Guide to Sequence-Based Identification of Biodiversity: An Interview with Simon Creer (2017) (https://bit.ly/2Cpb8Ty).
Podcast BioPod: Co-producer and presenter of BioPOD, the official podcast of the School of Natural Sciences at the University of Edinburgh.School Demonstrations: Demonstrator of Bite Size Biological practicals to school children.
Doors Open Days: The doors to the University are open and we get the opportunity to interact with the public about what we really get up to in our labs. This is a wonderful opportunity to engage with all people of all ages and backgrounds about what we do, and why we love science.
For the love of Science: Presented a poster on the science that I do and love at the public event engagement ‘for the love of science’ at the University of Exeter
Research
Current research
Title
PollerGEN - Using molecular genetics to understand grass species pollen deposition: enhancing bio-aerosol models and implications for human health.

Aims
Here a group of multidisciplinary researchers specialising in aerobiological modelling, DNA barcoding/molecular genetic identification and environmental health have teamed up with the UK Met Office in order to:
- Develop a species level, spatio-temporal grass pollen assessment framework throughout the UK
- Develop novel pollen bio-aerosol models
- Identify which species, or combinations of species are linked to asthma exacerbation
Summary
In Europe and the UK, the single most important outdoor aeroallergen is grass pollen, however grasses are particularly difficult to distinguish due to few unique morphological features. This project is in collaboration with the MET office who currently monitor pollen counts in the UK to provide a pollen forecast. However, there are currently no atmospheric models which successfully describe or predict atmospheric concentrations of grass pollen species. This is partly because there is no data on the presence and abundance of vegetation and pollen at the species level. The current level of discrimination of grass pollen is at the family level.
Moreover, the methods currently employed have not changed since WWII; the observations network is not resilient or financially sustainable and the forecast output is heavily reliant upon the skills, experience and longevity of individual observers. DNA-based identification has the potential to reduce processing time, produce methodologies which are easily shared between researchers and increase the level of species discrimination – and this is very important for grass pollen since different species of grass pollen cannot be discriminated using currently employed approaches of morphological examination under microscopy
Molecular genetic solutions
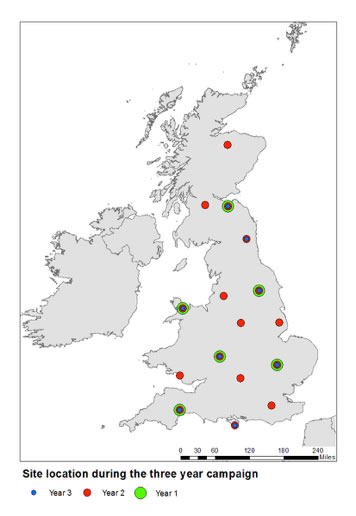
Pollen will be collected at up to 16 sites across the United Kingdom, over three annual cycles using volumetric methods.
- shotgun ultra-barcode (UBC) sequencing will be used to identify abundant grass pollen species in UK.
- qPCR will be used to quantify the abundances and temporal variation of up to 15 priority UK grass species.
- Illumina metabarcoding, will be used to measure the qualitative occurrences of the remaining PCR-amplifiable UK angiosperms.
Team PollerGEN is a group of multidisciplinary researchers

Lead investigator
Simon Creer (Bangor University)
Co-investigators of project
- Georgina Brennan (Bangor University)
- Natasha De Vere (Aberystwyth University/National Botanic Garden of Wales)
- Gareth Griffith (Aberystwyth University)
- Matthew Hegarty (Aberystwyth University)
- Carsten Skjøth (University of Worcester)
- Nicholas Osborne (University of Exeter/ University of Sydney)
- Ben Wheeler (University of Exeter)
- Adam Barber (Met Office)
- Yolanda Clewlow (Met Office)
- Rachel McInnes (Met Office)
Publications
2024
- PublishedAirborne DNA reveals predictable spatial and seasonal dynamics of fungi
Abrego, N., Furneaux, B., Hardwick, B., Somervuo, P., Palorinne, I., Aguilar-Trigueros, C. A., Andrew, N. R., Babiy, U. V., Bao, T., Bazzano, G., Bondarchuk, S. N., Bonebrake, T. C., Brennan, G. L., Bret-Harte, S., Bässler, C., Cagnolo, L., Cameron, E. K., Chapurlat, E., Creer, S., D'Acqui, L. P., de Vere, N., Desprez-Loustau, M-L., Dongmo, M. A. K., Jacobsen, I. B. D., Fisher, B. L., Flores de Jesus, M., Gilbert, G. S., Griffith, G. W., Gritsuk, A. A., Gross, A., Grudd, H., Halme, P., Hanna, R., Hansen, J., Hansen, L. H., Hegbe, A. D. M. T., Hill, S., Hogg, I. D., Hultman, J., Hyde, K. D., Hynson, N. A., Ivanova, N., Karisto, P., Kerdraon, D., Knorre, A., Krisai-Greilhuber, I., Kurhinen, J., Kuzmina, M., Lecomte, N., Lecomte, E., Loaiza, V., Lundin, E., Meire, A., Mešić, A., Miettinen, O., Monkhouse, N., Mortimer, P., Müller, J., Nilsson, R. H., Nonti, P. Y. C., Nordén, J., Nordén, B., Norros, V., Paz, C., Pellikka, P., Pereira, D., Petch, G., Pitkänen, J-M., Popa, F., Potter, C., Purhonen, J., Pätsi, S., Rafiq, A., Raharinjanahary, D., Rakos, N., Rathnayaka, A. R., Raundrup, K., Rebriev, Y. A., Rikkinen, J., Rogers, H. M. K., Rogovsky, A., Rozhkov, Y., Runnel, K., Saarto, A., Savchenko, A., Schlegel, M., Schmidt, N. M., Seibold, S., Skjøth, C., Stengel, E., Sutyrina, S. V., Syvänperä, I., Tedersoo, L., Timm, J., Tipton, L., Toju, H., Uscka-Perzanowska, M., van der Bank, M., van der Bank, F. H., Vandenbrink, B., Ventura, S., Vignisson, S. R., Wang, X., Weisser, W. W., Wijesinghe, S. N., Wright, S. J., Yang, C., Yorou, N. S., Young, A., Yu, D. W., Zakharov, E. V., Hebert, P. D. N., Roslin, T. & Ovaskainen, O., 25 Jul 2024, In: Nature. 631, 8022, p. 835-842 8 p.
Research output: Contribution to journal › Article › peer-review
2021
- PublishedPredicting the severity of the grass pollen season and the effect of climate change in Northwest Europe
Kurganskiy, A., Creer, S., De Vere, N., Griffith, G. W., Osborne, N. J., Wheeler, B. W., McInnes, R. N., Clewlow, Y., Barber, A., Brennan, G., Hanlon, H. M., Hegarty, M. J., Potter, C., Rowney, F. M., Adams-Groom, B., Petch, G. M., Pashley, C. H., Satchwell, J., De Weger, L. A., Rasmussen, K., Oliver, G., Sindt, C., Bruffaerts, N., The PollerGEN Consortium & Skjoth, C. A., 26 Mar 2021, In: Science Advances. 7, 13, eabd7658.
Research output: Contribution to journal › Article › peer-review
2020
- PublishedKey Questions for Next-Generation Biomonitoring
Makiola, A., Compson, Z. G., Baird, D. J., Barnes, M. A., Boerlijst, S. P., Bouchez, A., Brennan, G., Bush, A., Canard, E., Cordier, T., Creer, S., Curry, R. A., David, P., Dumbrell, A. J., Gravel, D., Hajibabaei, M., Hayden, B., van der Hoorn, B., Jarne, P., Jones, J. I., Karimi, B., Keck, F., Kelly, M., Knot, I. E., Krol, L., Massol, F., Monk, W. A., Murphy, J., Pawlowski, J., Poisot, T., Porter, T. M., Randall, K. C., Ransome, E., Ravigne, V., Raybould, A., Robin, S., Scrama, M., Schatz, B., Tamaddoni-Nezhad, A., Trimbos, K. B., Vacher, C., Vasselon, V., Wood, S., Woodward, G. & Bohan, D. A., 9 Jan 2020, In: Frontiers in Environmental Science.
Research output: Contribution to journal › Article › peer-review
2019
- PublishedOcean acidification increases iodine accumulation in kelp-based coastal food webs: Ocean acidification increases iodine in kelp
Xu, D., Brennan, G., Xu, L., Zhang, X. W., Fan, X., Han, W. T., Mock, T., McMinn, A., Hutchins, D. A. & Ye, N., Feb 2019, In: Global Change Biology. 25, 2, p. 629-639
Research output: Contribution to journal › Article › peer-review - PublishedTemperate airborne grass pollen defined by spatio-temporal shifts in community composition
Brennan, G. L., Potter, C., De Vere, N., Griffith, G. W., Skjøth, C. A., Osborne, N. J., Wheeler, B. W., McInnes, R. N., Clewlow, Y., Barber, A., Hanlon, H. M., Hegarty, M., Jones, L., Kurganskiy, A., Rowney, F. M., Armitage, C., Adams-Groom, B., Ford, C. R., Petch, G. M., The PollerGEN Consortium & Creer, S., May 2019, In: Nature Ecology and Evolution. 3, 5, p. 750-754
Research output: Contribution to journal › Article › peer-review
2017
- PublishedEvolutionary consequences of multidriver environmental change in an aquatic primary producer
Brennan, G., Colegrave, N. & Collins, S., 12 Sept 2017, In: Proceedings of the National Academy of Sciences of the USA. 114, 37, p. 9930–9935
Research output: Contribution to journal › Article › peer-review
2015
- PublishedGrowth responses of a green alga to multiple environmental drivers
Brennan, G. & Collins, S., Sept 2015, In: Nature Climate Change. 5, p. 892-899 7 p.
Research output: Contribution to journal › Article › peer-review
2014
- PublishedUnderstanding macroalgal dispersal in a complex hydrodynamic environment: a combined population genetic and physical modelling approach
Brennan, G., Kregting, L., Beatty, G., Cole, C., Elsäßer, B., Savidge, G. & Provan, J., 26 Mar 2014, In: Journal of the Royal Society, Interface. 11, 95, p. 1 12 p.
Research output: Contribution to journal › Article › peer-review

