Professor Si Creer

- Name
- Professor Si Creer
- Position
- Professor of Molecular Ecology
- s.creer@bangor.ac.uk
- Phone
- +44 1248 382302
- Location
- School of Natural Sciences, Bangor University, Deiniol Road, Bangor, Gwynedd, LL57 2UW, UK
About
I am interested in using contemporary molecular tools to address diverse questions focusing on biodiversity, ecology and evolution. This is a particularly exciting time in the field of molecular ecology, since advances in DNA sequencing throughput have recently offered a paradigm shift in our ability to assess previously intractable functional and taxonomic biodiversity at an unprecedented scale, augmenting existing biodiversity fields and empowering others. Using such technologies, I am testing a range of hypotheses regarding the alpha and beta functional and taxonomic diversity of macro-, meio- and microbial communities (e.g. microbiomes) in space and time, based on genomic, community and environmental DNA (eDNA). Focal habitats have included estuarine, coastal and deep sea environments with an increasing focus now on freshwater, terrestrial, whole organisms and the aerial biosphere in order to understand the drivers of diversity in natural communities and also how diversity is linked with ecological function, trophic relationships, environmental and human health. Current additional activities include phylogenomics, population genetics, life history evolution, polyploidy, pollination genomics. If you want to find out more, please go to my research and publication pages.
If you are interested in this sort of work or life, please follow me on Twitter: @spideycreer 
Recent Methods in Ecology and Evolution podcast on the use of DNA sequencing to identify biodiversity (Creer et al. MEE 2016) also available here.
CV
Education
- 2005 Teaching in Higher Education Certificate. UWB.
- 2000 PhD. Molecular phylogeography and venom evolution of Trimeresurus stejnegeri in Taiwan. NERC funded, The University of Wales, Bangor (UWB), UK.
- 1997 MSc. Ecology (Distinction). NERC funded, UWB.
- 1992 BSc. (Hons) Applied Biology (2.1). University of Bath, UK.
Career
- 9/16 – Professor of Molecular Ecology – Bangor University.
- 11/12–9/16 – Senior Lecturer in Molecular Ecology – Bangor University.
- 11/05–10/11 Senior Research Fellow in Molecular Ecology – Bangor University.
- 07/05–10/05 Postdoctoral Research Associate. NERC grant, “Functional and genomic venom diversity of phospholipase A2 (PLA2) genes in Asian pitvipers” UWB.
- 06/01–06/05 Postdoctoral Research Associate (named). Wellcome Trust grant, “Reliable molecular phylogenies for medically important venomous snakes: subtropical and tropical Asian vipers” UWB.
- 09/03–09/04 Temporary Lectureship in Zoology. UWB.
- 01–06/01 Postdoctoral Research Associate. Leverhulme Trust grant, “Far-east Asian pitvipers: venom evolution and biodiversity” University of Wales, Bangor.
Grants and Awards
- 2016–2019 NERC Standard Grant (PI) – PollerGEN – Using molecular genetics to understand grass species pollen deposition: enhancing bio-aerosol models and implications for human health (£1.2M, with Natasha DeVere, Gareth Griffith, Matt Hegarty, Aberystwyth; Carsten Skjøth, Worcester; Nick Osborne and Ben Wheeler, Exeter and the UK Met Office)
- 2015–2020 NERC Highlight Topic Grant (PI) – Understanding the ecological relevance of eDNA in freshwater lotic ecosystems (LOFRESH: £1.25M, with Mark DeBruyn, Gary Carvalho; CEH Bangor; John Colbourne, Holly Bik, Birmingham and Isabelle Durance and Steve Ormerod, Cardiff)
- 2015–2020 NERC Large Grant (CoI) – Impacts of global warming in sentinel systems: from genes to ecosystems (£3.7M, with Guy Woodward, Imperial)
- 2015–2017 Marie Curie Intra-European Fellowship (CoI) with Laura Kelly – PARMIN (£200K)
- 2014–2016 Royalty Research Fund Small Grant, University of Washington (CoI) with Lorenz Hauser – Tackling the meiofauna paradox with metagenetics ($40K)
- 2014–2016 Formas, Sweden (CoI) – 2 year mobility grant with Francisco Nascimento (£150K)
- 2016–2017 Vetenskapsrådet (VR) (CoI) One year postdoc grant with Francisco Nascimento (£31K)
- 2014–2015 SciLifeLab (CoI) – National projects in Swedish Genomes and Biodiversity with Francisco Nascimento (£15K)
- 2013–2016 Welsh Government Grant (CoI) – Glastir Monitoring and Evaluation Programme (GMEP): Soil biodiversity (£4M, with CEH and GMEP Consortium)
- 2013–2015 AQUATRACE: Linking life-history traits and genetic variation (Co-I) in collaboration with Einar Nielsen DTU-Aqua Denmark (168K Euros)
- 2012–2015 – FISHPROBIO (PI) : Marie Curie Outgoing Fellowship with Martin Llewelyn (290,000 Euros)
- 2011–2013 National Science Centre (Poland) (CoI) – Seasonal differences in diversity of bacteria and pico- and nanoplanktonic protists in 3 zones of the Vistula River estuary (£121K)
- 2010–2011 – NERC AFI (PI) Collaborative Gearing Scheme grant: A second-generation sequencing perspective of the Antarctic bentho-pelagic microbial biosphere
- 2010–2012 – MARMEDIV (PI) : Marie Curie Incoming Fellowship with Frederic Sinniger (174,000 Euros)
- 2009 - Natural Environment Research Council (NERC) (PI) Environmental genomics of life in the intertidal: Design and optimization of a gastropod microarray (£8000)
- 2008–2010- NERC (PI)Post genomic and proteomics grant: Sequencing the meiofaunal metagenome of the marine/freshwater interface in key estuarine ecosystems (£140000)
- 2008- CoSyst (PI) : The Systematics Association, The Linnean Society, Biotechnology and Biological Sciences Research Council (BBSRC), NERC: Advancing mitogenomics via ultrasequencing: A case study within the Araneae (£10200)
- 2007 – NERC (PI) Phylochip development for the rapid identification of littoral meiofaunal communities (£4750)
- 2006–2009 – NERC (PI) New Investigator Award (PI) : Are marine nematodes hyperdiverse? A metagenomic solution (£76000)
- 2004 Joseph B. Slowinski Award for the most distinguished international paper on snake systematics for Creer et al (2003a) ($500)
- 1997 UWB Llewellyn and Mary Williams Scholarship for PhD funding (£14000)
- 1996 NERC MSc funding (£3500)
Professional Activities
- 2019- Subject Editor Ecography - Ecography
- 2018- Advisory Board eDNA
- 2017- present Member of the Genetics Society
- 2016-2018 Fellow of the Royal Society of Biology (FRSB)
- 2013- Associate Editor Molecular Ecology
- 2010- Associate Editor Molecular Ecology Resources
- 2010 - present Member of the British Ecological Society
- 2009 - 2018 Member of the Systematics Association
- 2009- NERC Peer Review College member
- 2006–2009 Committee member for the Joseph B. Slowinski Award
- 2005 Instigator of UW Bangor becoming a member organisation of the Consortium for Barcoding of Life
- 2003-present Member of the British Arachnological Society (BAS)
In addition to the above, I review articles for Nature Communications, Systematic Biology, Molecular Ecology, Philosophical Transactions and Proceedings of the Royal Society, Biological Sciences, BMC Evolutionary Biology, Conservation Genetics, Heredity, Journal of Molecular Evolution, Journal of Experimental Marine Biology and Ecology, Journal of Fish Biology, Journal of Zoology, Environmental Microbiology and Marine Ecology Progress Series.
Research
Research interests
LOFRESH – Understanding the ecological relevance of lotic environmental DNA (eDNA)
In recent years, paradigm shifts have been made in using DNA sourced directly from environmental samples (eDNA), to identify large-scale patterns of biodiversity from taxa across the tree of life. LOFRESH is a multi-institution collaborative initiative funded by a Natural Environment Research Council (NERC) Highlight Topic grant to improve our ability to utilize eDNA for tracking the presence and abundance of species in and around freshwater habitats. Environmental DNA refers to shed cells or extracellular DNA from organisms as they pass through an environment (e.g. water, soil and air), or die and decay. By doing so, animals and plants leave traces of their DNA in the environment that can be detected using a number of molecular genetic approaches.
We expect that our findings will provide valuable insights for the fields of freshwater ecology, biomonitoring and environmental assessment. Launched in March 2016, LOFRESH aims to understand the dynamics between living communities and lotic (i.e. riverine) eDNA in relation to spatial and environmental variation.
While led by Bangor University, LOFRESH is also directly supported by The Centre for Ecology and Hydrology (CEH), Cardiff University and the University of Birmingham. Please visit the project website for further details and please do get in touch if you would like to explore future synergies with the project, or associated research areas.
NERC PollerGEN – Understanding links between grass pollen and human health in the UK
For millions of people, the onset of spring and summer brings misery as they battle with itchy eyes and sneezing brought about by their reaction to pollen. Up to 25% of the UK population are sensitised to grass pollen and around 10% suffer asthma that can be aggravated by pollen. However, with around 150 different species of grass in the UK and no easy way of distinguishing between different pollen grains, identifying which species of grass pollen people are allergic to is a challenge. 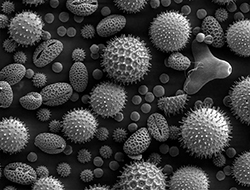
In PollerGEN, a £1.2M NERC investment, we aim to develop a species level, spatio-temporal pollen assessment framework throughout the UK and develop novel pollen bio-aerosol models in order to identify which species are linked to the exacerbation of asthma.
The PollerGEN team comprises researchers from Bangor, Aberystwyth, Worcester and Exeter Universities, in collaboration with the UK Met office, supported by a range of stakeholder groups and charities. We will use a range of molecular genetic solutions, from shotgun environmental sequencing through to quantitative PCR to quantify the pollen landscape, followed by leading-edge aerobiological modelling and identifying links to the aggravation of asthma throughout the grass pollen season.
Understanding which species of grass pollens are in the air in high quantities at a particular time will allow those with hay fever and asthma to better manage their disease by being aware of risky periods, avoiding exposure and having their medicines to hand. It is predicted that the new forecasts will give more precise (and shorter) time periods that disease sufferers will have to be cautious in, and provide guidance of when these may occur.
Click here to view the Past Research Projects.
Publications
2024
- PublishedA diversity of diversities: Do complex environmental effects underpin associations between below- and above-ground taxa?
Seaton, F., George, P., Alison, J., Jones, D. L., Creer, S., Smart, S. M., Emmett, B. A. & Robinson, D., 1 Jul 2024, In: Journal of Ecology. 112, 7, p. 1550-1564 15 p.
Research output: Contribution to journal › Article › peer-review - PublishedAirborne DNA reveals predictable spatial and seasonal dynamics of fungi
Abrego, N., Furneaux, B., Hardwick, B., Somervuo, P., Palorinne, I., Aguilar-Trigueros, C. A., Andrew, N. R., Babiy, U. V., Bao, T., Bazzano, G., Bondarchuk, S. N., Bonebrake, T. C., Brennan, G. L., Bret-Harte, S., Bässler, C., Cagnolo, L., Cameron, E. K., Chapurlat, E., Creer, S., D'Acqui, L. P., de Vere, N., Desprez-Loustau, M-L., Dongmo, M. A. K., Jacobsen, I. B. D., Fisher, B. L., Flores de Jesus, M., Gilbert, G. S., Griffith, G. W., Gritsuk, A. A., Gross, A., Grudd, H., Halme, P., Hanna, R., Hansen, J., Hansen, L. H., Hegbe, A. D. M. T., Hill, S., Hogg, I. D., Hultman, J., Hyde, K. D., Hynson, N. A., Ivanova, N., Karisto, P., Kerdraon, D., Knorre, A., Krisai-Greilhuber, I., Kurhinen, J., Kuzmina, M., Lecomte, N., Lecomte, E., Loaiza, V., Lundin, E., Meire, A., Mešić, A., Miettinen, O., Monkhouse, N., Mortimer, P., Müller, J., Nilsson, R. H., Nonti, P. Y. C., Nordén, J., Nordén, B., Norros, V., Paz, C., Pellikka, P., Pereira, D., Petch, G., Pitkänen, J-M., Popa, F., Potter, C., Purhonen, J., Pätsi, S., Rafiq, A., Raharinjanahary, D., Rakos, N., Rathnayaka, A. R., Raundrup, K., Rebriev, Y. A., Rikkinen, J., Rogers, H. M. K., Rogovsky, A., Rozhkov, Y., Runnel, K., Saarto, A., Savchenko, A., Schlegel, M., Schmidt, N. M., Seibold, S., Skjøth, C., Stengel, E., Sutyrina, S. V., Syvänperä, I., Tedersoo, L., Timm, J., Tipton, L., Toju, H., Uscka-Perzanowska, M., van der Bank, M., van der Bank, F. H., Vandenbrink, B., Ventura, S., Vignisson, S. R., Wang, X., Weisser, W. W., Wijesinghe, S. N., Wright, S. J., Yang, C., Yorou, N. S., Young, A., Yu, D. W., Zakharov, E. V., Hebert, P. D. N., Roslin, T. & Ovaskainen, O., 25 Jul 2024, In: Nature. 631, 8022, p. 835-842 8 p.
Research output: Contribution to journal › Article › peer-review - PublishedAn integrated spatio-temporal view of riverine biodiversity using environmental DNA metabarcoding
Perry, W. B., Seymour, M., Orsini, L., Jâms, I. B., Milner, N., Edwards, F., Harvey, R., de Bruyn, M., Bista, I., Walsh, K., Emmett, B., Blackman, R., Altermatt, F., Lawson Handley, L., Mächler, E., Deiner, K., Bik, H. M., Carvalho, G., Colbourne, J., Cosby, B. J., Durance, I. & Creer, S., 23 May 2024, In: Nature Communications. 15, 1, p. 4372
Research output: Contribution to journal › Article › peer-review - PublishedBacillus indicus and Bacillus subtilis as alternative health and colouration promoters to synthetic astaxanthin in cyprinid aquaculture species.
Baumgärtner, S., Creer, S., Jones, C., James, J. & Ellison, A., 15 Jan 2024, In: Aquaculture. 578, 740016.
Research output: Contribution to journal › Article › peer-review - PublishedDesign considerations for eDNA metabarcoding surveys
Perry, W. B., Pillay, K., George, P., Brennan, G., Lowe, A., Jones, L., Holman, L., Gibson, T., de Vere, N. & Creer, S., 19 Jan 2024, Applied Environmental Genomics. Berry, O. F., Holleley, C. E. & Jarman, S. N. (eds.). CRC Press
Research output: Chapter in Book/Report/Conference proceeding › Chapter › peer-review - E-pub ahead of printEnvironmental DNA reveals ecologically relevant spatial and temporal variation in fish assemblages between estuaries and seasons
Gibson, T., Baillie, C., Collins, R. A., Wangensteen, O. S., Corrigan, L., Ellison, A., Heddell-Cowie, M., Westoby, H., Byatt, B., Lawson-Handley, L., Soto, A. Z., Creer, S., Genner, M. J. & Mariani, S., 1 Aug 2024, In: Ecological Indicators. 165, 112215.
Research output: Contribution to journal › Article › peer-review - PublishedMolecular ecology of microbiomes in the wild: Common pitfalls, methodological advances and future directions
Fountain-Jones, N. M., Giraud, T., Zinger, L., Bik, H., Creer, S. & Videvall, E., 12 Jan 2024, In: Molecular Ecology. 33, 2, e1723.
Research output: Contribution to journal › Article › peer-review
2023
- Published100 years of anthropogenic impact causes changes in freshwater functional biodiversity
Eastwood, N., Zhou, J., Derelle, R., Abdallah, M. A-E., Stubbings, W. A., Jia, Y., Crawford, S. E., Davidson, T. A., Colbourne, J. K., Creer, S., Bik, H., Hollert, H. & Orsini, L., 7 Nov 2023, In: Elife. 12
Research output: Contribution to journal › Article › peer-review - E-pub ahead of printComparative population genomics of manta rays has global implications for management
Humble, E., Hosegood, J., Carvalho, G., De Bruyn, M., Creer, S., Stevens, G. M. W., Armstrong, A., Bonfil, R., Deakos, M., Fernando, D., Froman, N., Peel, L. R., Pollett, S., Ponzo, A., Stewart, J. D., Wintner, S. & Ogden, R., 23 Nov 2023, (E-pub ahead of print) In: Molecular Ecology.
Research output: Contribution to journal › Article › peer-review - PublishedEnvironmental DNA biomonitoring in biodiversity hotspots: A case study of fishes of the Okavango Delta
von der Heyden, S., Neef, G., Grevesse, T., Cwecwe, Y., Sado, T., Miya, M., Mosie, I., Creer, S., Skelton, P. & von Brandis, R., 28 Nov 2023, In: Environmental DNA. 5, 6, p. 1720-1731
Research output: Contribution to journal › Article › peer-review - PublishedEnvironmental DNA metabarcoding for fish diversity assessment in a macrotidal estuary: A comparison with established fish survey methods
Gibson, T., Carvalho, G., Ellison, A., Gargiulo, E., Hatton-Ellis, T., Handley, L. L., Mariani, S., Collins, R. A., Sellers, G., Distaso, M., Zampieri, C. & Creer, S., 1 Nov 2023, In: Estuarine, Coastal and Shelf Science. 294, 108522.
Research output: Contribution to journal › Article › peer-review - PublishedMicroscale pollen release and dispersal patterns in flowering grass populations
Frisk, C. A., Apangu, G. P., Petch, G. M., Creer, S., Hanson, M., Adams-Groom, B. & Skjoth, C. A., 1 Jul 2023, In: Science of the Total Environment. 880, 163345.
Research output: Contribution to journal › Article › peer-review - PublishedMineralogy affects prokaryotic community composition in an acidic metal mine
Kelly, L., Rivett, D. W., Pakostova, E., Creer, S., Cotterell, T. & Johnson, B., 1 Jan 2023, In: Microbiological Research. 266, 127257.
Research output: Contribution to journal › Article › peer-review - PublishedUK DNA working group eDNA week, January 2022
Handley, L. L., Blackwell, T., Broadhurst, H. A., Clark, K., Davison, P. I., England, J., Mariani, S., McDevitt, A. D., Pillay, G. K., Read, D., Walsh, K., Nisbet, A. & Creer, S., 1 Jan 2023, In: Environmental DNA. 5, 1, p. 18-24 7 p.
Research output: Contribution to journal › Article › peer-review
2022
- PublishedChallenges to Implementing Environmental-DNA Monitoring in Namibia
Perry, I., Jams, I. B., Casas-Mulet, R., Hamutoko, J., Marchbank, A., Lendelvo, S., Naomab, E., Mapani, B., Creer, S., Wanke, H., Durance, I. & Kille, P., 17 Jan 2022, In: Frontiers in Environmental Science. 9, 773991.
Research output: Contribution to journal › Article › peer-review - PublishedFauxcurrence: simulating multi-species occurrences for null models in species distribution modelling and biogeography
Osborne, O., Fell, H. G., Atkins, H., Tol, J. V., Phillips, D., Herrerra-Alsina, L., Mynard, P., Bocedi, G., Gubry-Rangin, C., Lancaster, L. T., Creer, S., Nangoy, M., Fahri, F., Lupiyaningdyah, P., Sudiana, I. M., Juliandi, B., Travis, J., Papadopulos, A. S. T. & Algar, A. C., Jul 2022, In: Ecography. 2022, 7, e05880.
Research output: Contribution to journal › Article › peer-review - PublishedLong term drought and warming alter soil bacterial and fungal communities in an upland heathland
Seaton, F., Reinsch, S., Goodall, T., White, N., Jones, D. L., Griffiths, R., Creer, S., Smith, A., Emmett, B. A. & Robinson, D. A., Sept 2022, In: Ecosystems. 25, 6, p. 1279-1294 16 p.
Research output: Contribution to journal › Article › peer-review - PublishedManaging human-mediated range shifts: understanding spatial, temporal and genetic variation in marine non-native species
Holman, L. E., Parker-Nance, S., de Bruyn, M., Creer, S., Carvalho, G. & Rius, M., 14 Mar 2022, In: Philosophical Transactions of The Royal Society B: Biological Sciences. 377, 1846, 20210025.
Research output: Contribution to journal › Article › peer-review - PublishedNovel insights into marine fish biodiversity across a pronounced environmental gradient using replicated environmental DNA analyses
Czachur, M. V., Seymour, M., Creer, S. & von der Heyden, S., 1 Jan 2022, In: Environmental DNA. 4, 1
Research output: Contribution to journal › Article › peer-review - PublishedSeasonal progression and differences in major floral resource use by bees and hoverflies in a diverse horticultural and agricultural landscape revealed by DNA metabarcoding
Lowe, A., Jones, L., Brennan, G., Creer, S. & de Vere, N., Jun 2022, In: Journal of Applied Ecology. 59, 6, p. 1484-1495
Research output: Contribution to journal › Article › peer-review - PublishedStrategies for sample labelling and library preparation in DNA metabarcoding studies
Bohmann, K., Elbrecht, V., Caroe, C., Bista, I., Leese, F., Bunce, M., Yu, D. W., Seymour, M., Dumbrell, A. & Creer, S., May 2022, In: Molecular Ecology Resources. 22, 4, p. 1231-1246 16 p.
Research output: Contribution to journal › Review article › peer-review - PublishedTemporal Patterns of Honeybee Foraging in a Diverse Floral Landscape Revealed Using Pollen DNA Metabarcoding of Honey
Jones, L., Lowe, A., Ford, C. R., Creer, S. & de Vere, N., 1 Aug 2022, In: Integrative and Comparative Biology. 62, 2, p. 199-210
Research output: Contribution to journal › Article › peer-review - PublishedTemporal change in floral availability leads to periods of resource limitation and affects diet specificity in a generalist pollinator
Lowe, A., Jones, L., Brennan, G., Creer, S., Christie, L. & de Vere, N., 24 Oct 2022, In: Molecular Ecology. 32, 23, p. 6363-6376 14 p.
Research output: Contribution to journal › Article › peer-review - PublishedUsing DNA Metabarcoding to Identify Floral Visitation by Pollinators
Lowe, A., Jones, L., Witter, L., Creer, S. & de Vere, N., 24 Mar 2022, In: Diversity. 14, 4
Research output: Contribution to journal › Review article › peer-review
2021
- PublishedAncient geological dynamics impact neutral biodiversity accumulation and are detectable in phylogenetic reconstructions
Herrera‐Alsina, L., Algar, A. C., Bocedi, G., Gubry‐Rangin, C., Lancaster, L., Mynard, P., Osborne, O., Papadopulos, A. S. T., Creer, S., Nangoy, M., Fahri, F., Lupiyaningdyah, P., Sudiana, I. M., Juliandi, B. & Travis, J. M. J., Aug 2021, In: Global Ecology and Biogeography. 30, 8, p. 1633-1642 10 p.
Research output: Contribution to journal › Article › peer-review - PublishedAnimals, protists and bacteria share marine biogeographic patterns
Holman, L. E., De Bruyn, M., Creer, S., Carvalho, G., Robidart, J. & Rius, M., 1 Jun 2021, In: Nature Ecology and Evolution. 5, 6, p. 738–746
Research output: Contribution to journal › Article › peer-review - PublishedConnecting high‐throughput biodiversity inventories: Opportunities for a site‐based genomic framework for global integration and synthesis
Arribas, P., Andujar, C., Bidartondo, M. I., Bohmann, K., Coissac, E., Creer, S., deWaard, J. R., Elbrecht, V., Ficetola, G. F., Goberna, M., Kennedy, S., Krehenwinkel, H., Leese, F., Novotny, V., Ronquist, F., Yu, D. W., Zinger, L., Creedy, T. J., Meramveliotakis, E., Noguerales, V., Overcast, I., Morlon, H., Vogler, A. P., Papadopoulou, A. & Emerson, B. C., 1 Mar 2021, In: Molecular Ecology. 30, 5, p. 1120-1135 16 p.
Research output: Contribution to journal › Article › peer-review - PublishedDNA Metabarcoding Methods for the Study of Marine Benthic Meiofauna
Gielings, R., Fais, M., Fontaneto, D., Creer, S., Costa, F. O., Renema, W. & Macher, J-N., 30 Sept 2021, In: Frontiers in Marine Science.
Research output: Contribution to journal › Article › peer-review - PublishedDecoupled richness of generalist anaerobes and sulphate-reducing bacteria is driven bypHacross land uses in temperate soils
George, P. B. L., Coelho, K. P., Creer, S., Lebron, I., Robinson, D. A. & Jones, D. L., Nov 2021, In: European Journal of Soil Science. 72, 6, p. 2445-2456 12 p.
Research output: Contribution to journal › Article › peer-review - PublishedDomestication-induced reduction in eye size revealed in multiple common garden experiments: The case of Atlantic salmon (Salmo salar L.)
Perry, W. B., Kaufmann, J., Solberg, M. F., Brodie, C., Medina, A. M. C., Pillay, K., Egerton, A., Harvey, A., Phillips, K. P., Coughlan, J., Egan, F., Grealis, R., Hutton, S., Leseur, F., Ryan, S., Poole, R., Rogan, G., Ryder, E., Schaal, P., Waters, C., Wynne, R., Taylor, M., Prodohl, P., Creer, S., Llewellyn, M., McGinnity, P., Carvalho, G. & Glover, K. A., 28 Sept 2021, In: Evolutionary Applications. 14, 9, p. 2319-2332 14 p.
Research output: Contribution to journal › Article › peer-review - PublishedEcosystems monitoring powered by environmental genomics: a review of current strategies with an implementation roadmap
Cordier, T., Alonso-Saez, L., Apothéloz-Perret-Gentil, Aylagas, E., Bohan, D. A., Bouchez, A., Chariton, A. A., Creer, S., Fruhe, L., Keck, F., Keeley, N., Laroche, O., Leese, F., Pochon, X., Stoeck, T., Pawlowski, J. & Lanzen, A., Jul 2021, In: Molecular Ecology. 30, 13, p. 2937-2958 22 p.
Research output: Contribution to journal › Article › peer-review - PublishedEnvironmental DNA monitoring of oncogenic viral shedding and genomic profiling of sea turtle fibropapillomatosis reveals unusual viral dynamics
Farrell, J. A., Yetsko, K., Whitmore, L., Whilde, J., Eastman, C. B., Ramia, D. R., Thomas, R., Linser, P., Creer, S., Burkhalter, B., Schnitzler, C. & Duffy, D. J., 12 May 2021, In: Communications Biology. 4, 1, p. 565 1 p.
Research output: Contribution to journal › Article › peer-review - PublishedEnvironmental DNA provides higher resolution assessment of riverine biodiversity and ecosystem function via spatio-temporal nestedness and turnover partitioning
Seymour, M., Edwards, F. K., Cosby, B. J., Bista, I., Scarlett, P. M., Brailsford, F., Glanville, H., de Bruyn, M., Carvalho, G. & Creer, S., 3 May 2021, In: Communications Biology. 4, 1, 512.
Research output: Contribution to journal › Article › peer-review - PublishedEnvironmental DNA reveals links between abundance and composition of airborne grass pollen and respiratory health
Rowney, F. M., Brennan, G. L., Skjoth, C. A., Griffith, G. W., McInnes, R. N., Clewlow, Y., Adams-Groom, B., Barber, A., De Vere, N., Economou, T., Hegarty, M., Hanlon, H. M., Jones, L., Kurganskiy, A., Petch, G. M., Potter, C., Munawar Rafiq, A., Warner, A., The PollerGEN Consortium, Wheeler, B. W., Osborne, N. J. & Creer, S., 10 May 2021, In: Current Biology. 31, 9, p. 1995-2003 9 p.
Research output: Contribution to journal › Article › peer-review - PublishedHigh throughput shotgun sequencing of eRNA reveals taxonomic and derived functional shifts across a benthic productivity gradient
Broman, E., Bonaglia, S., Norkko, A., Creer, S. & Nascimento, F. J. A., Jul 2021, In: Molecular Ecology. 30, 13, p. 3023-3039 17 p.
Research output: Contribution to journal › Article › peer-review - PublishedMolecular characterization of a marine turtle tumor epizootic, profiling external, internal and postsurgical regrowth tumors
Yetsko, K., Farrell, J., Blackburn, N. B., Whitmore, L., Stammnitz, M. R., Whilde, J., Eastman, C., Ramia, D. R., Thomas, R., Krstic, A., Linser, P., Creer, S., Carvalho, G., Devlin, M. A., Nahvi, N., Leandro, A. C., deMaar, T. W., Burkhalter, B., Murchison, E. P., Schnitzler, C. & Duffy, D., 1 Feb 2021, In: Communications Biology. 4, 1, 152.
Research output: Contribution to journal › Article › peer-review - PublishedPredicting the severity of the grass pollen season and the effect of climate change in Northwest Europe
Kurganskiy, A., Creer, S., De Vere, N., Griffith, G. W., Osborne, N. J., Wheeler, B. W., McInnes, R. N., Clewlow, Y., Barber, A., Brennan, G., Hanlon, H. M., Hegarty, M. J., Potter, C., Rowney, F. M., Adams-Groom, B., Petch, G. M., Pashley, C. H., Satchwell, J., De Weger, L. A., Rasmussen, K., Oliver, G., Sindt, C., Bruffaerts, N., The PollerGEN Consortium & Skjoth, C. A., 26 Mar 2021, In: Science Advances. 7, 13, eabd7658.
Research output: Contribution to journal › Article › peer-review - PublishedShifts in Soil Structure, Biological, and Functional Diversity Under Long-Term Carbon Deprivation
George, P. B. L., Fidler, D. B., Van Nostrand, J. D., Atkinson, J. A., Mooney, S. J., Creer, S., Griffiths, R. I., McDonald, J. E., Robinson, D. A. & Jones, D. L., 14 Sept 2021, In: Frontiers in Microbiology. 12, 735022.
Research output: Contribution to journal › Article › peer-review - PublishedShifts in honeybee foraging reveal historical changes in floral resources
Jones, L., Brennan, G. L., Lowe, A., Creer, S., Ford, C. R. & de Vere, N., 14 Jan 2021, In: Communications Biology. 4, 1, 37.
Research output: Contribution to journal › Article › peer-review - PublishedSoil health cluster analysis based on national monitoring of soil indicators
Seaton, F. M., Barrett, G., Burden, A., Creer, S., Fitos, E., Garbutt, A., Griffiths, R. I., Henrys, P., Jones, D. L., Keenan, P., Keith, A., Lebron, I., Maskell, L., Pereira, M. G., Reinsch, S., Smart, S. M., Williams, B., Emmett, B. A. & Robinson, D. A., Nov 2021, In: European Journal of Soil Science. 72, 6, p. 2414-2429 16 p.
Research output: Contribution to journal › Article › peer-review
2020
- PublishedChapter Ten - Informing marine spatial planning decisions with environmental DNA
Bani, A., De Brauwer, M., Creer, S., Dumbrell, A. J., Limmon, G., Jompa, J., von der Heyden, S. & Berger, M., 2020, In: Advances in Ecological Research. 62, p. 375-407
Research output: Contribution to journal › Article › peer-review - PublishedDisentangling the effects of sex, life history and genetic background in Atlantic salmon: growth, heart and liver under common garden conditions
Perry, W. B., Solberg, M. F., Brodie, C., Medina, A. C., Pillay, K. G., Egerton, A., Harvey, A., Creer, S., Llewellyn, M., Taylor, M., Carvalho, G. & Glover, K. A., 7 Oct 2020, In: Royal Society Open Science. 7, 10, 200811.
Research output: Contribution to journal › Article › peer-review - PublishedEpistatic regulation of growth in Atlantic salmon revealed: a QTL study performed on the domesticated-wild interface
Besnier, F., Solberg, M. F., Harvey, A., Carvalho, G., Bekkevold, D., Taylor, M. I., Creer, S., Nielsen, E. E., Skaala, O., Ayllon, F., Dahle, G. & Glover, K. A., 7 Feb 2020, In: BMC Genetics. 21, 1, 13.
Research output: Contribution to journal › Article › peer-review - PublishedExecuting multi-taxa eDNA ecological assessment via traditional metrics and interactive networks
Seymour, M., Edwards, F., Cosby, B., Kelly, M., de Bruyn, M., Carvalho, G. & Creer, S., 10 Aug 2020, In: Science of the Total Environment. 729, 138801.
Research output: Contribution to journal › Article › peer-review - PublishedKey Questions for Next-Generation Biomonitoring
Makiola, A., Compson, Z. G., Baird, D. J., Barnes, M. A., Boerlijst, S. P., Bouchez, A., Brennan, G., Bush, A., Canard, E., Cordier, T., Creer, S., Curry, R. A., David, P., Dumbrell, A. J., Gravel, D., Hajibabaei, M., Hayden, B., van der Hoorn, B., Jarne, P., Jones, J. I., Karimi, B., Keck, F., Kelly, M., Knot, I. E., Krol, L., Massol, F., Monk, W. A., Murphy, J., Pawlowski, J., Poisot, T., Porter, T. M., Randall, K. C., Ransome, E., Ravigne, V., Raybould, A., Robin, S., Scrama, M., Schatz, B., Tamaddoni-Nezhad, A., Trimbos, K. B., Vacher, C., Vasselon, V., Wood, S., Woodward, G. & Bohan, D. A., 9 Jan 2020, In: Frontiers in Environmental Science.
Research output: Contribution to journal › Article › peer-review - PublishedPhylogenomics and species delimitation for effective conservation of manta and devil rays
Hosegood, J., Humble, E., Ogden, R., De Bruyn, M., Creer, S., Stevens, G. M. W., Abudaya, M., Bassos-Hull, K., Bonfil, R., Fernando, D., Foote, A., Hipperson, H., Jabado, R. W., Kaden, J., Moazzam, M., Peel, L. R., Pollett, S., Ponzo, A., Poortvliet, M., Salah, J., Senn, H., Stewart, J. D., Wintner, S. & Carvalho, G., Dec 2020, In: Molecular Ecology. 29, 24, p. 4783-4796
Research output: Contribution to journal › Article › peer-review - PublishedSoil textural heterogeneity impacts bacterial but not fungal diversity
Seaton, F., George, P., Lebron, I., Jones, D. L., Creer, S. & Robinson, D. A., 1 May 2020, In: Soil Biology and Biochemistry. 144, 107766.
Research output: Contribution to journal › Article › peer-review
2019
- PublishedAbove-below surface interactions mediate effects of seagrass disturbance on meiobenthic diversity, nematode and polychaete trophic structure
Nascimento, F. J. A., Dahl, M., Deyanova, D., Lyimo, L. D., Bik, H. M., Pereira, T. J., Bjork, M., Creer, S. & Gullstrom, M., 4 Oct 2019, In: Communications Biology. 2, 362.
Research output: Contribution to journal › Article › peer-review - PublishedCryptic diets of forage fish: jellyfish consumption observed in the Celtic Sea and western English Channel
Lamb, P. D., Hunter, E., Pinnegar, J. K., van der Kooij, J., Creer, S. & Taylor, M. I., 26 Jun 2019, In: Journal of Fish Biology. 94, 6, p. 1026-1032 7 p.
Research output: Contribution to journal › Article › peer-review - PublishedDNA metabarcoding-Need for robust experimental designs to draw sound ecological conclusions
Zinger, L., Bonin, A., Alsos, I. G., Bálint, M., Bik, H., Boyer, F., Chariton, A. A., Creer, S., Coissac, E., Deagle, B. E., De Barba, M., Dickie, I. A., Dumbrell, A. J., Ficetola, G. F., Fierer, N., Fumagalli, L., Gilbert, M. T. P., Jarman, S., Jumpponen, A., Kauserud, H., Orlando, L., Pansu, J., Pawlowski, J., Tedersoo, L., Thomsen, P. F., Willerslev, E. & Taberlet, P., Apr 2019, In: Molecular Ecology. 28, 8, p. 1857-1862 6 p.
Research output: Contribution to journal › Article › peer-review - PublishedDeep segregation in the open ocean: Macaronesia as an evolutionary hotspot for low dispersal marine invertebrates
Vieira, P. E., Desiderato, A., Holdich, D. M., Soares, P., Creer, S., Carvalho, G., Costa, F. O. & Queiroga, H., 1 Apr 2019, In: Molecular Ecology. 28, 7, p. 1784-1800
Research output: Contribution to journal › Article › peer-review - PublishedDetection of introduced and resident marine species using environmental DNA metabarcoding of sediment and water
Holman, L. E., de Bruyn, M., Creer, S., Carvalho, G., Robidart, J. & Rius, M., 9 Aug 2019, In: Scientific Reports. 9, 11559.
Research output: Contribution to journal › Article › peer-review - PublishedDivergent national-scale trends of microbial and animal biodiversity revealed across diverse temperate soil ecosystems
George, P., Lallias, D., Creer, S., Seaton, F., Kenny, J. G., Eccles, R. M., Griffiths, R., Lebron, I., Emmett, B., Robinson, D. & Jones, D. L., 7 Mar 2019, In: Nature Communications. 10, 1, p. 1107 1107.
Research output: Contribution to journal › Article › peer-review - PublishedEnvironmental DNA size sorting and degradation experiment indicates the state of Daphnia magna mitochondrial and nuclear eDNA is subcellular
Moushomi, R., Wilgar, G., Carvalho, G., Creer, S. & Seymour, M., 29 Aug 2019, In: Scientific Reports. 9, 12500 (2019.
Research output: Contribution to journal › Article › peer-review - PublishedEvolutionary drivers of kype size in Atlantic salmon (Salmo salar): domestication, age and genetics
Perry, W. B., Solberg, M. F., Besnier, F., Dyrhovden, L., Matre, I. H., Fjelldal, P. G., Ayllon, F., Creer, S., Llewelyn, M., Taylor, M. I., Carvalho, G. & Glover, K. A., 17 Apr 2019, In: Royal Society Open Science. 6, 4, 190021.
Research output: Contribution to journal › Article › peer-review - PublishedHow quantitative is metabarcoding: a meta-analytical approach
Lamb, P. D., Hunter, E., Pinnegar, J. K., Creer, S., Taylor, M. I. & Davies, R. G., Jan 2019, In: Molecular Ecology. 28, 2, p. 420-430 11 p.
Research output: Contribution to journal › Article › peer-review - PublishedInclusion of jellyfish in 30+ years of Ecopath with Ecosim models
Lamb, P. D., Hunter, E., Pinnegar, J. K., Doyle, T. K., Creer, S. & Taylor, M. I., 1 Dec 2019, In: ICES Journal of Marine Science. 76, 7, p. 1941–1950
Research output: Contribution to journal › Article › peer-review - PublishedIntroduction: Special issue on species interactions, ecological networks and community dynamics – Untangling the entangled bank using molecular techniques
Roslin, T., Traugott, M., Jonsson, M., Stone, G. N., Creer, S. & Symondson, W. O. C., 1 Jan 2019, In: Molecular Ecology. 28, 2, p. 157-164
Research output: Contribution to journal › Article › peer-review - PublishedPlant and soil communities are associated with the response of soil water repellency to environmental stress
Seaton, F., Jones, D. L., Creer, S., George, P., Smart, S. M., Lebron, I., Barrett, G., Emmett, B. & Robinson, D., 15 Oct 2019, In: Science of the Total Environment. 687, p. 929-938
Research output: Contribution to journal › Article › peer-review - PublishedPrimer and database choice affect fungal functional but not biological diversity findings in a national soil survey
George, P., Creer, S., Griffiths, R. I., Emmett, B. A., Robinson, D. A. & Jones, D. L., 1 Nov 2019, In: Frontiers in Environmental Science. 7, 173.
Research output: Contribution to journal › Article › peer-review - PublishedSalinity drives meiofaunal community structure dynamics across the Baltic ecosystem
Broman, E., Raymond, C., Sommer, C., Gunnarsson, J. S., Creer, S. & Nascimento, F. J. A., Aug 2019, In: Molecular Ecology. 28, 16, p. 3813-3829 17 p.
Research output: Contribution to journal › Article › peer-review - PublishedTemperate airborne grass pollen defined by spatio-temporal shifts in community composition
Brennan, G. L., Potter, C., De Vere, N., Griffith, G. W., Skjøth, C. A., Osborne, N. J., Wheeler, B. W., McInnes, R. N., Clewlow, Y., Barber, A., Hanlon, H. M., Hegarty, M., Jones, L., Kurganskiy, A., Rowney, F. M., Armitage, C., Adams-Groom, B., Ford, C. R., Petch, G. M., The PollerGEN Consortium & Creer, S., May 2019, In: Nature Ecology and Evolution. 3, 5, p. 750-754
Research output: Contribution to journal › Article › peer-review
2018
- PublishedAcidity promotes degradation of multi-species environmental DNA in lotic mesocosms
Seymour, M., Durance, I., Cosby, B. J., Ransom-Jones, E., Deiner, K., Ormerod, S. J., Colbourne, J. K., Wilgar, G., Carvalho, G., De Bruyn, M., Edwards, F., Emmett, B. A., Bik, H. M. & Creer, S., 22 Jan 2018, In: Communications Biology. 1, 4.
Research output: Contribution to journal › Article › peer-review - PublishedDetecting macroecological patterns in bacterial communities across independent studies of global soils
Ramirez, K. S., Knight, C. G., Hollander, M., Brearley, F. Q., Constantinides, B., Cotton, T. E. A., Creer, S., Crowther, T. W., Davison, J., Delgado-Baquerizo, M., Dorrepaal, E., Elliott, D. R., Fox, G., Griffiths, R., Hale, C., Hartman, K., Houlden, A., Jones, D., Krab, E. J., Maestre, F. T., McGuire, K. L., Monteux, S., Orr, C. H., van der Putten, W. H., Roberts, I. S., Robinson, D., Rocca, J., Rowntree, J., Schlaeppi, K., Shepherd, M., Singh, B. K., Straathof, A., Talbot, J. M., Thion, C., van der Heijden, M. & de Vries, F. T., 2018, In: Nature Microbiology. 3, p. 189-196 8 p.
Research output: Contribution to journal › Article › peer-review - PublishedDiversity and community composition of pico- and nanoplanktonic protists in the Vistula River estuary (Gulf of Gdańsk, Baltic Sea)
Piwosz, K., Calkiewicz, J., Golebiewski, M. & Creer, S., 31 Jul 2018, In: Estuarine, Coastal and Shelf Science. 207, p. 242-249
Research output: Contribution to journal › Article › peer-review - PublishedPerformance of amplicon and shotgun sequencing for accurate biomass estimation in invertebrate community samples
Bista, I., Carvalho, G., Tang, M., Walsh, K., Zhou, X., Hajibabaei, M., Shokralla, S., Seymour, M., Bradley, D., Liu, S., Christmas, M. & Creer, S., Sept 2018, In: Molecular Ecology Resources. 18, 5, p. 1020-1034
Research output: Contribution to journal › Article › peer-review - PublishedSample size effects on the assessment of eukaryotic diversity and community structure in aquatic sediments using high-throughput sequencing
Nascimento, F. J. A., Lallias, D., Bik, H. M. & Creer, S., 6 Aug 2018, In: Scientific Reports. 8
Research output: Contribution to journal › Article › peer-review - PublishedWhole genome duplication and transposable element proliferation drive genome expansion in Corydoradinae catfishes
Marburger, S., Alexandrou, M., Taggart, J. B., Creer, S., Carvalho, G., Oliveira, C. & Taylor, M. I., 14 Feb 2018, In: Proceedings of the Royal Society B: Biological Sciences. 285, 1872, 20172732.
Research output: Contribution to journal › Article › peer-review
2017
- PublishedA communal catalogue reveals Earth's multiscale microbial diversity
Thompson, L. R., Sanders, J. G., McDonald, D., Amir, A., Ladau, J., Locey, K. J., Prill, R. J., Tripathi, A., Gibbons, S. M., Ackermann, G., Navas-Molina, J. A., Janssen, S., Kopylova, E., Vazquez-Baeza, Y., Gonzalez, A., Morton, J. T., Mirarab, S., Xu, Z. Z., Jiang, L., Haroon, M. F., Kanbar, J., Zhu, Q., Song, S. J., Kosciolek, T., Bokulich, N. A., Lefler, J., Brislawn, C. J., Humphrey, G., Owens, S. M., Hampton-Marcell, J., Berg-Lyons, D., McKenzie, V., Fierer, N., Fuhrman, J. A., Clauset, A., Stevens, R. L., Shade, A., Pollard, K. S., Goodwin, K. D., Jansson, J. K., Gilbert, J. A., Knight, R., Agosto Rivera, J. L., Al-Moosawi, L., Alverdy, J., Amato, K. R., Andras, J., Angenent, L. T., Antonopoulos, D. A., Apprill, A., Armitage, D., Ballantine, K., Bárta, J., Baum, J. K., Berry, A., Bhatnagar, A., Bhatnagar, M., Biddle, J. F., Bittner, L., Boldgiv, B., Bottos, E., Boyer, D. M., Braun, J., Brazelton, W., Brearley, F. Q., Campbell, A. H., Caporaso, J. G., Cardona, C., Carroll, J., Cary, S. C., Casper, B. B., Charles, T. C., Chu, H., Claar, D. C., Clark, R. G., Clayton, J. B., Clemente, J. C., Cochran, A., Coleman, M. L., Collins, G., Colwel, R. R., Contreras, M., Crary, B. B., Creer, S., Cristol, D. A., Crump, B. C., Cui, D., Daly, S. E., Davalos, L., Dawson, R. D., Defazio, J., Delsuc, F., Dionisi, H. M., Dominguez-Bello, M. G., Dowell, R., Dubinsky, E. A., Dunn, P. O., Ercolini, D., Espinoza, R. E., Ezenwa, V., Fenner, N., Findlay, H. S., Fleming, I. D., Fogliano, V., Forsman, A., Freeman, C., Friedman, E. S., Galindo, G., Garcia, L., Garcia-Amado, M. A., Garshelis, D., Gasser, R. B., Gerdts, G., Gibson, M. K., Gifford, I., Gill, R. T., Giray, T., Gittel, A., Golyshin, P., Gong, D., Grossart, H-P., Guyton, K., Haig, S-J., Hale, V., Hall, R. S., Hallam, S. J., Handley, K. M., Hasan, N. A., Haydon, S. R., Hickman, J. E., Hidalgo, G., Hofmockel, K. S., Hooker, J., Hulth, S., Hultman, J., Hyde, E., Ibáñez-Álamo, J. D., Jastrow, J. D., Jex, A. R., Johnson, L. S., Johnston, E. R., Joseph, S., Jurburg, S. D., Jurelevicius, D., Karlsson, A., Karlsson, R., Kauppinen, S., Kellogg, C. T. E., Kennedy, S. J., Kerkhof, L. J., King, G. M., Kling, G. W., Koehler, A. V., Krezalek, M., Kueneman, J., Lamendella, R., Landon, E. M., Lane-deGraaf, K., LaRoche, J., Larsen, P., Laverock, B., Lax, S., Lentino, M., Levin, I. I., Liancourt, P., Liang, W., Linz, A. M., Lipson, D. A., Liu, Y., Lladser, M. E., Lozada, M., Spirito, C. M., MacCormack, W. P., MacRae-Crerar, A., Magris, M., Martín-Platero, A. M., Martín-Vivaldi, M., Martínez, L. M., Martínez-Bueno, M., Marzinelli, E. M., Mason, O. U., Mayer, G. D., McDevitt-Irwin, J. M., McDonald, J., McGuire, K. L., McMahon, K. D., McMinds, R., Medina, M., Mendelson, J. R., Metcalf, J. L., Meyer, F., Michelangeli, F., Miller, K., Mills, D. A., Minich, J., Mocali, S., Moitinho-Silva, L., Moore, A., Morgan-Kiss, R. M., Munroe, P., Myrold, D., Neufeld, J. D., Ni, Y., Nicol, G. W., Nielsen, S., Nissimov, J. I., Niu, K., Nolan, M. J., Noyce, K., O’Brien, S. L., Okamoto, N., Orlando, L., Ortiz Castellano, Y., Osuolale, O., Oswald, W., Parnell, J., Peralta-Sánchez, J. M., Petraitis, P., Pfister, C., Pilon-Smits, E., Piombino, P., Pointing, S. B., Pollock, F. J., Potter, C., Prithiviraj, B., Quince, C., Rani, A., Ranjan, R., Rao, S., Rees, A. P., Richardson, M., Riebesell, U., Robinson, C., Rockne, K. J., Rodriguezl, S. M., Rohwer, F., Roundstone, W., Safran, R. J., Sangwan, N., Sanz, V., Schrenk, M., Schrenzel, M. D., Scott, N. M., Seger, R. L., Seguin-Orlando, A., Seldin, L., Seyler, L. M., Shakhsheer, B., Sheets, G. M., Shen, C., Shi, Y., Shin, H., Shogan, B. D., Shutler, D., Siegel, J., Simmons, S., Sjöling, S., Smith, D. P., Soler, J. J., Sperling, M., Steinberg, P. D., Stephens, B., Stevens, M. A., Taghavi, S., Tai, V., Tait, K., Tan, C. L., Tas, N., Taylor, D. L., Thomas, T., Timling, I., Turner, B. L., Urich, T., Ursell, L. K., van der Lelie, D., Van Treuren, W., van Zwieten, L., Vargas-Robles, D., Vega Thurber, R., Vitaglione, P., Walker, D. A., Walters, W. A., Wang, S., Wang, T., Weave, T., Webster, N. S., Wehrle, B., Weisenhorn, P., Weiss, S., Werner, J. J., West, K., Whitehead, A., Whitehead, S. R., Whittingham, L. A., Willerslev, E., Williams, A. E., Wood, S. A., Woodhams, D. C., Yang, Y., Zaneveld, J., Zarraonaindia, I., Zhang, Q. & Zhao, H., 23 Nov 2017, In: Nature. 551, 7681, p. 457-+
Research output: Contribution to journal › Article › peer-review - PublishedAnnual time-series analysis of aqueous eDNA reveals ecologically relevant dynamics of lake ecosystem biodiversity
Bista, I., Carvalho, G., Walsh, K., Seymour, M., Hajibabaei, M., Lallias, D., Christmas, M. & Creer, S., Jan 2017, In: Nature Communications. 8, 14087.
Research output: Contribution to journal › Article › peer-review - PublishedEnvironmental DNA metabarcoding: transforming how we survey animal and plant communities
Deiner, K., Bik, H. M., Machler, E., Seymour, M., Lacoursiere-Roussel, A., Altermatt, F., Creer, S., Bista, I. & Lodge, D. M., Nov 2017, In: Molecular Ecology Resources. 26, 21, p. 5872-5895
Research output: Contribution to journal › Article › peer-review - PublishedEvaluation of mesofauna communities as soil quality indicators in a national-level monitoring programme
George, P., Keith, A. M., Creer, S., Barrett, G. L., Lebron, I., Emmett, B. A., Robinson, D. & Jones, D., 1 Dec 2017, In: Soil Biology and Biochemistry. 115, p. 537-546
Research output: Contribution to journal › Article › peer-review - PublishedJellyfish on the menu: mtaDNA assay reveals scyphozoan predation in the Irish Sea
Lamb, P. D., Hunter, E., Pinnegar, J. K., Creer, S., Davies, R. G. & Taylor, M. I., 29 Nov 2017, In: Royal Society Open Science.
Research output: Contribution to journal › Article › peer-review - PublishedMarine ecology: Genetics from a drop in the ocean
Creer, S. & Seymour, M., 4 Jan 2017, In: Nature Ecology and Evolution. 1, 0037.
Research output: Contribution to journal › Comment/debate - PublishedParasitism perturbs the mucosal microbiome of Atlantic Salmon
Llewellyn, M. S., Leadbeater, S., Garcia, C., Sylvain, F-E., Custodio, M., Ang, K. P., Powell, F., Carvalho, G., Creer, S., Elliot, J. & Derome, N., 2017, In: Scientific Reports. 7, 43465.
Research output: Contribution to journal › Article › peer-review - PublishedRevealing higher than expected meiofaunal diversity in Antarctic sediments: a metabarcoding approach
Fonseca, V. G., Sinninger, F., Gaspar, J. M., Quince, C., Creer, S., Power, D., Peck, L. S. & Clark, M. S., 21 Jul 2017, In: Scientific Reports. 7, 6094.
Research output: Contribution to journal › Article › peer-review - PublishedSubtle shifts in microbial communities occur alongside the release of carbon induced by drought and rewetting in contrasting peatland ecosystems
Potter, C., Freeman, C., Golyshin, P., Ackerman, G., Fenner, N., McDonald, J., Ehbair, A., Jones, T., Murphy, L. & Creer, S., Sept 2017, In: Scientific Reports. 7, 11314.
Research output: Contribution to journal › Article › peer-review - PublishedTideless estuaries in brackish seas as possible freshwater-marine transition zones for bacteria: the case study of the Vistula river estuary
Golebiewski, M., Calkiewicz, J., Creer, S. & Piwosz, K., Apr 2017, In: Environmental Microbiology Reports. 9, 2, p. 129-143
Research output: Contribution to journal › Article › peer-review - PublishedUsing DNA metabarcoding to investigate honey bee foraging reveals limited flower use despite high floral availability
De Vere, N., Jones, L. E., Gilmore, T., Moscrop, J., Lowe, A., Smith, D., Hegarty, M. J., Creer, S. & Ford, C. R., 2017, In: Scientific Reports. 7, 42838.
Research output: Contribution to journal › Article › peer-review
2016
- PublishedA common garden design reveals population-specific variability in potential impacts of hybridization between populations of farmed and wild Atlantic salmon, Salmo salar L.
Harvey, A., Glover, K. A., Taylor, M., Creer, S. & Carvalho, G., 1 Mar 2016, In: Evolutionary Applications. 9, 3, p. 435-449
Research output: Contribution to journal › Article › peer-review - PublishedCoexisting cryptic species of the Litoditis marina complex (Nematoda) show differential resource use and have distinct microbiomes with high intraspecific variability
Derycke, S., De Meester, N., Rigaux, A., Creer, S., Bik, H. M., Thomas, W. K. & Moens, T., May 2016, In: Molecular Ecology. 25, 9, p. 2093-2110
Research output: Contribution to journal › Article › peer-review - PublishedDoes density influence relative growth performance of farmed, wild, and F1 hybrid Atlantic salmon in semi-natural and hatchery common garden conditions?
Harvey, A., Carvalho, G., Taylor, M., Solberg, M. F., Creer, S., Dyrhovden, L., Matre, I. H. & Glover, K. A., 6 Jul 2016, In: Royal Society Open Science. 3, 7, 160152.
Research output: Contribution to journal › Article › peer-review - PublishedPlasticity in growth of farmed and wild Atlantic salmon: is the increased growth rate of farmed salmon caused by evolutionary adaptations to the commercial diet?
Harvey, A., Solberg, M. F., Troianou, E., Carvalho, G., Taylor, M. I., Creer, S., Dyrhovden, L., Matre, I. H. & Glover, K. A., 1 Dec 2016, In: BMC Evolutionary Biology. 2016, 16, 264.
Research output: Contribution to journal › Article › peer-review - PublishedPlasticity in response to feed availability: Does feeding regime influence the relative growth performance of domesticated, wild and hybrid Atlantic salmon Salmo salar parr?
Harvey, A., Solberg, M. F., Glover, K. A., Taylor, M. I., Creer, S. & Carvalho, G., 1 Sept 2016, In: Journal of Fish Biology. 89, 3, p. 1754-1768
Research output: Contribution to journal › Article › peer-review - PublishedThe biogeography of the atlantic salmon (Salmo salar) gut microbiome
Llewellyn, M. S., McGinnity, P., Dionne, M., Letourneau, J., Thonier, F., Carvalho, G., Creer, S. & Derome, N., May 2016, In: ISME Journal. 10, p. 1280-1284
Research output: Contribution to journal › Article › peer-review - PublishedThe ecologist’s field guide to sequence-based identification of biodiversity
Creer, S., Deiner, K., Frey, S., Porazinska, D., Taberlet, P., Thomas, W. K., Potter, C. & Bik, H. M., 13 Sept 2016, In: Methods in Ecology and Evolution. 7, p. 1008-1018 11 p.
Research output: Contribution to journal › Article › peer-review - PublishedWorldwide Analysis of Sedimentary DNA Reveals Major Gaps in Taxonomic Knowledge of Deep-Sea Benthos
Sinniger, F., Pawlowski, J., Harii, S., Gooday, A. J., Yamamoto, H., Chevaldonne, P., Cedhagen, T., Carvalho, G. & Creer, S., 14 Jun 2016, In: Frontiers in Marine Science. 3, 92
Research output: Contribution to journal › Article › peer-review
2015
- PublishedConvergence of multiple markers and analysis methods defines the genetic distinctiveness of cryptic pitvipers
Mrinalini, M., Thorpe, R. S., Creer, S., Lallias, D. S., Dawnay, L., Stuart, B. L. & Malhotra, A., 8 Jul 2015, In: Molecular Phylogenetics and Evolution. 92, p. 266-279
Research output: Contribution to journal › Article › peer-review - PublishedThe importance of being genomic: Non-coding and coding sequences suggest different models of toxin multi-gene family evolution
Malhotra, A., Creer, S., Harris, J. B. & Thorpe, R. S., 7 Sept 2015, In: Toxicon. 107, Part B, p. 344-358
Research output: Contribution to journal › Article › peer-review
2014
- PublishedA critique of Rossberg et al.: noise obscures the genetic signal of meiobiotal ecospecies in ecogenomic datasets
Morgan, M. J., Bass, D., Bik, H., Birky, C. W., Blaxter, M., Crisp, M. D., Derycke, S., Fitch, D., Fonteaneto, D., Hardy, C. M., King, A. J., Kiontke, K. C., Moens, T., Pawlowski, J. W., Porazinska, D., Tang, C. Q., Thomas, W. K., Yeates, D. K. & Creer, S., 26 Mar 2014, In: Proceedings of the Royal Society B: Biological Sciences. 281, 1783, p. 20133076
Research output: Contribution to journal › Article › peer-review - PublishedEnvironmental DNA for Wildlife Biology and Biodiversity Monitoring
Evans, A. R., Bohmann, K., Evans, A., Gilbert, M. T., Carvalho, G. R., Creer, S., Knapp, M., Yu, D. W. & De Bruyn, M., 10 May 2014, In: Trends in Ecology and Evolution. 29, 6, p. 358-367
Research output: Contribution to journal › Article › peer-review - PublishedEnvironmental metabarcoding reveals heterogeneous drivers of microbial eukaryote diversity in contrasting estuarine ecosystems
Lallias, D. S., Lallias, D., Hiddink, J. G., Fonseca, V. G., Gaspar, J. M., Sung, W., Neill, S. P., Barnes, N., Ferrero, T., Hall, N., Lambshead, P. J., Packer, M., Thomas, W. K. & Creer, S., 25 Nov 2014, In: The ISME Journal. 9, p. 1208-1221
Research output: Contribution to journal › Article › peer-review - PublishedMetagenetic analysis of patterns of distribution and diversity of marine meiobenthic eukaryotes
Fonseca, V. G., Carvalho, G. R., Nichols, B., Quince, C., Johnson, H. F., Neill, S. P., Lambshead, J. D., Thomas, W. K., Power, D. M. & Creer, S., 25 Aug 2014, In: Global Ecology and Biogeography. 23, 11, p. 1293–1302
Research output: Contribution to journal › Article › peer-review
2013
- PublishedCan Long-Range PCR Be Used to Amplify Genetically Divergent Mitochondrial Genomes for Comparative Phylogenetics? A Case Study within Spiders (Arthropoda: Araneae)
Briscoe, A. G., Goodacre, S., Masta, S. E., Taylor, M. I., Arnedo, M. A., Penney, D., Kenny, J. & Creer, S., 8 May 2013, In: PLoS ONE. 8, 5, p. e62404
Research output: Contribution to journal › Article › peer-review - PublishedExperimental harvesting of fish populations drives genetically based shifts in body size and maturation
Van Wijk, S. J., van Wijk, S. J., Taylor, M. I., Creer, S., Dreyer, C., Rodrigues, F. M., Ramnarine, I. W., van Oosterhout, C. & Carvalho, G. R., 1 May 2013, In: Frontiers in Ecology and the Environment. 11, 4, p. 181–187
Research output: Contribution to journal › Article › peer-review - PublishedInvestigating the molecular systematic relationships amongst selected Plesionika (Decapoda: Pandalidae) from the Northeast Atlantic and Mediterranean Sea
da Silva, J. M., dos Santos, A., Cunha, M. R., Costa, F. O., Creer, S. & Carvalho, G. R., 1 Jun 2013, In: Marine Ecology. 34, 2, p. 157-170
Research output: Contribution to journal › Article › peer-review - PublishedPredicting function from sequence in a large multifunctional toxin family
Malhotra, A., Creer, S., Harris, J. B., Stöcklin, R., Favereau, P. & Thorpe, R. S., 3 Jul 2013, In: Toxicon. 72, p. 113-125
Research output: Contribution to journal › Article › peer-review - PublishedSNP Discovery in European Anchovy (Engraulis encrasicolus, L) by High-Throughput Transcriptome and Genome Sequencing
Montes, I., Conklin, D., Albania, A., Creer, S., Carvalho, G. R., Santos, M. & Estonba, A., 1 Aug 2013, In: PLoS ONE. 8, 8, p. e70051
Research output: Contribution to journal › Article › peer-review - PublishedTranscriptomics and in vivo tests reveal novel mechanisms underlying endocrine disruption in an ecological sentinel, Nucella lapillus
Hughes, R. N., Pascoal, S., Carvalho, G. R., Vasieva, O., Hughes, R., Cossins, A., Fang, Y., Ashelford, K., Olohan, L., Barroso, C., Mendo, S. & Creer, S., 1 Mar 2013, In: Molecular Ecology. 22, 6, p. 1589-1608
Research output: Contribution to journal › Article › peer-review
2012
- PublishedCosmopolitanism of microbial eukaryotes in the global deep seas
Creer, S. & Sinniger, F., 1 Mar 2012, In: Molecular Ecology. 21, 5, p. 1033-1035
Research output: Contribution to journal › Article › peer-review - PublishedPlastic and Heritable Components of Phenotypic Variation in Nucella lapillus: An Assessment Using Reciprocal Transplant and Common Garden Experiments
Hughes, R. N., Pascoal, S., Carvalho, G. R., Creer, S., Rock, J., Kawaii, K., Mendo, S. & Hughes, R., 27 Jan 2012, In: PLoS ONE. 7, 1, p. e30289
Research output: Contribution to journal › Article › peer-review - PublishedSample richness and genetic diversity as drivers of chimera formation in nSSU metagenetic analyses
Fonseca, V. G., Nichols, B., Lallias, D., Quince, C., Carvalho, G. R., Power, D. M. & Creer, S., 25 Jan 2012, In: Nucleic Acids Research. 40, 9, p. e66
Research output: Contribution to journal › Article › peer-review - PublishedSequencing our way towards understanding global eukaryotic biodiversity
Bik, H. M., Porazinska, D., Creer, S., Caporaso, J. G., Knight, R. & Thomas, W. K., 1 Apr 2012, In: Trends in Ecology and Evolution. 27, 4, p. 233-243
Research output: Contribution to journal › Article › peer-review
2011
- PublishedCompetition and phylogeny determine community structure in Müllerian co-mimics
Alexandrou, M. A., Oliveira, C., Maillard, M., McGill, R. A., Newton, J., Creer, S. & Taylor, M. I., 1 Jun 2011, In: Nature. 469, p. 84-88
Research output: Contribution to journal › Article › peer-review - PublishedTemporal genetic homogeneity among shore crab (Carcinus maenas) larval events supplied to an estuarine system on the Portuguese northwest coast.
Domingues, C. P., Creer, S., Taylor, M. I., Queiroga, H. & Carvalho, G. R., 1 May 2011, In: Heredity. 106, 5, p. 832-840
Research output: Contribution to journal › Article › peer-review
2010
- PublishedGenetic structure of Carcinus maenas within its native range: larval dispersal and oceanographic variability.
Domingues, C. P., Creer, S., Taylor, M. I., Queiroga, H. & Carvalho, G. R., 14 Jul 2010, In: Marine Ecology Progress Series. 410, p. 111-123
Research output: Contribution to journal › Article › peer-review - PublishedInclusion of nuclear intron sequence data helps to identify the Asian sister group of New World pitvipers.
Malhotra, A., Creer, S., Pook, C. E. & Thorpe, R. S., 1 Jan 2010, In: Molecular Phylogenetics and Evolution. 54, 1, p. 172-178
Research output: Contribution to journal › Article › peer-review - PublishedSecond-generation environmental sequencing unmasks marine metazoan biodiversity.
Fonseca, V. G., Carvalho, G. R., Sung, W., Johnson, H. F., Power, D. M., Neill, S. P., Packer, M., Blaxter, M. L., Lambshead, J. D., Thomas, W. K. & Creer, S., 19 Oct 2010, In: Nature Communications. 1, 7, p. 98
Research output: Contribution to journal › Article › peer-review - PublishedSecond-generation sequencing derived insights into the temporal biodiversity dynamics of freshwater protists.
Creer, S., 1 Jul 2010, In: Molecular Ecology. 19, 14, p. 2829-2831
Research output: Contribution to journal › Article › peer-review - PublishedUltrasequencing of the meiofaunal biosphere: practice, pitfalls and promises.
Creer, S., Fonseca, V. G., Porazinska, D. L., Giblin-Davis, R. M., Sung, W., Powers, D. M., Packer, M., Carvalho, G. R., Blaxter, M. L., Lambshead, P. J. & Thomas, W. K., 1 Mar 2010, In: Molecular Ecology. 19, 1, p. 4-20
Research output: Contribution to journal › Article › peer-review
2009
- PublishedAn evaluation of the systematic value of skull morphology in the Trimeresurus radiation (Serpentes: Viperidae: Crotalinae) of Asian pitvipers.
Guo, P., Malhotra, A., Creer, S. & Pook, C. E., 1 Nov 2009, In: Journal of Zoological Systematics and Evolutionary Research. 47, 4, p. 378-384
Research output: Contribution to journal › Article › peer-review - PublishedComments on the systematic status of specimens belonging to the genus Viridovipera (Serpentes: Viperidae: Crotalinae) from Sichuan and Yunnan provinces of southwestern China, with a redescription of V. yunnanensis.
Guo, P., Malhotra, A., Thorpe, R. S., Creer, S. & Pook, C. E., 1 Jul 2009, In: Herpetological Journal. 19, 3, p. 151-162
Research output: Contribution to journal › Article › peer-review - PublishedDevelopment and Application of Microsatellites in Carcinus maenas: Genetic Differentiation between Northern and Central Portuguese Populations
Pascoal, S., Creer, S., Taylor, M. I., Queiroga, H., Carvalho, G. & Mendo, S., 30 Sept 2009, In: PLoS ONE. 4, 9, p. e7268
Research output: Contribution to journal › Article › peer-review - PublishedSystematics of the Protobothrops jerdonii complex (Serpentes, Viperidae, Crotalinae) inferred from morphometric data and molecular phylogeny.
Guo, P., Malhotra, A., Li, C., Creer, S., Pook, P. E. & Wen, T., 1 Jul 2009, In: Herpetological Journal. 19, 2, p. 85-96
Research output: Contribution to journal › Article › peer-review
2007
- PublishedChoosing and using introns in molecular phylogenetics.
Creer, S., 1 Jan 2007, In: Evolutionary Bioinformatics. 3, p. 99-108
Research output: Contribution to journal › Article › peer-review - PublishedNew evidence on the phylogenetic position of the poorly known Asian pitviper Protobothrops kaulbacki (Serpentes : Viperidae : Crotalinae) with a redescription of the species and a revision of the genus Protobothrops.
Guo, P., Malhotra, A., Li, P. P., Pook, C. E. & Creer, S., 1 Oct 2007, In: Herpetological Journal. 17, 4, p. 237-246
Research output: Contribution to journal › Article › peer-review
2006
- PublishedOptimal intron analyses in the Trimeresurus radiation of Asian pitvipers.
Creer, S., Pook, C. E., Malhotra, A. & Thorpe, R. S., 1 Feb 2006, In: Systematic Biology. 55, 1, p. 57-72
Research output: Contribution to journal › Article › peer-review
2005
- PublishedOn the application of molecular barcodes in toxinological research
Creer, S., 1 Nov 2005, In: Toxicon. 46, 6, p. 709-710
Research output: Contribution to journal › Article › peer-review - PublishedTargeting optimal introns for phylogenetic analyses in non-model taxa: experimental results in Asian pitvipers.
Creer, S., Malhotra, A., Thorpe, R. S. & Pook, C. E., 1 Aug 2005, In: Cladistics. 21, 4, p. 390-395
Research output: Contribution to journal › Article › peer-review
2004
- PublishedThe utility of AFLPs for supporting mitochondrial DNA phylogeographical analyses in the Taiwanese bamboo viper, Trimeresurus stejnegeri
Creer, S., Thorpe, R., Malhotra, A., Chou, W. H. & Stenson, A. G., 1 Jan 2004, In: Journal of Evolutionary Biology. 17, 1, p. 100-107
Research output: Contribution to journal › Article › peer-review
2003
- PublishedAssessing the phylogenetic utility of four mitochondrial genes and a nuclear intron in the Asian pit viper genus, Trimeresurus: Separate, simultaneous, and conditional data combination analyses
Creer, S., Malhotra, A. & Thorpe, R. S., 1 Aug 2003, In: Molecular Biology and Evolution. 20, 8, p. 1240-1251
Research output: Contribution to journal › Article › peer-review - PublishedGenetic and ecological correlates of intraspecific variation in pitviper venom composition detected using matirx-assisted laser desorption time-of-flight mass spectrometry (MALDI-TOF-MS) and isoelectric focusing
Creer, S., Malhotra, A., Thorpe, R., Stocklin, R. S., Favreau, P. S. & Chou, W. S., 1 Mar 2003, In: Journal of Molecular Evolution. 56, 3, p. 317-329
Research output: Contribution to journal › Article › peer-review
2002
- PublishedOffshore insular variation in the diet of the Taiwanese bamboo viper Trimeresurus stejnegeri (Schmidt)
Creer, S., Chou, W. H., Malhotra, A. & Thorpe, R. S., 1 Aug 2002, In: Zoological Science. 19, 8, p. 907-913
Research output: Contribution to journal › Article › peer-review
2001
- PublishedMultiple causation of phylogeographical pattern as revealed by nested clade analysis of the bamboo viper (Trimeresurus stejnegeri) within Taiwan.
Creer, S., Malhotra, A., Thorpe, R. S. & Chou, W. H., 1 Aug 2001, In: Molecular Ecology. 10, 8, p. 1967-1981
Research output: Contribution to journal › Article › peer-review
