Staff Profile of Dr Nicholas Casewell

- Name
- Dr Nicholas Casewell
- Position
- NERC Research Fellow
- n.casewell@bangor.ac.uk
- Phone
- +44(0)1248 382318
- Location
- 3rd Floor, ECW Building, School of Biological Sciences, Bangor University, Deiniol Road, Bangor, Gwynedd, LL57 2UW, UK
About
I am interested in using multi-disciplinary tools to address questions relating to evolutionary biology; such as the role selection pressures play in trait evolution and how multi-locus gene families evolve.
To investigate these areas I have been using venomous animals as model organisms. Snake venoms provide a model system to assess extreme modes of evolution, with the gene families that encode venom components (toxins) typically evolving rapidly by frequent gene duplication and directional selection. I have been using molecular and evolutionary techniques to infer the evolutionary history of the venom ‘character’ and the gene families that encode venom toxins. In contrast to the predatory venoms used by snakes, a number of fish lineages contain defensive venoms that appear to have evolved independently.
I am currently using proteomic and molecular techniques to assess the composition of venom from a number of bony and cartilaginous fish lineages in order to reconstruct the evolutionary history of piscine venoms. As an aside from my interest in evolutionary biology, I am also involved in research projects focusing on improving the safety, stability and efficacy of snake antivenoms.
CV
Quick CV
Education
2010 PhD. “The genetic basis of venom variation in the genus Echis: causes, correlates and consequences”. Natural Environment Research Council (NERC) funded, Molecular Ecology and Evolution Group, Bangor University, UK.
2005 BSc (Hons). Tropical Disease Biology (1st). University of Liverpool, UK.
Career
09/12 – present NERC Postdoctoral Research Fellow. “Origin, evolution and functional diversification of a chemical arsenal: venom in body and cartilaginous fish”. Molecular Ecology and Evolution Group, Bangor University, UK
10/12 - 11/12 Visiting academic. Venom Evolution Laboratory, School of Biological Sciences, University of Queensland, Australia.
05/10 – 08/12 Postdoctoral Research Associate. “Design, development and testing of novel veterinary and human antivenoms”. Alistair Reid Venom Research Unit, Liverpool School of Tropical Medicine, UK and MicroPharm Limited, UK.
Grants and Awards
2012-2015 - NERC Postdoctoral Research Fellowship: “Origin, evolution and functional diversification of a chemical arsenal: venom in body and cartilaginous fish”. £293,000.
2012-2015 – Leverhume Trust grant: “Has defence dictated the evolution of venom composition in spitting cobras?” £209,000. Named Researcher.
2012 – Award for International Collaborative Research, University of Queensland, Australia.
2011 – Walter M. Fitch Award Finalist. Society for Molecular Biology and Evolution.
2011 – Walter M. Fitch conference travel award. Society for Molecular Biology and Evolution. £1,000.
2005 – Tropical Disease Biology Honours prize, School of Biological Sciences, University of Liverpool, UK.
2004 – W.P. Sinclair University Undergraduate Scholarship, University of Liverpool, UK.
Professional Activities
Keynote and invited speaker at ‘International Society on Toxinology’, ‘Venoms’, ‘Society for Molecular Biology and Evolution’ and ‘British Toxicological Society’ international conferences
Member of the International Society on Toxinology and the Society for Molecular Biology and Evolution
Article reviewer for Molecular Biology and Evolution, Molecular and Cellular Proteomics, Journal of Molecular Evolution, BMC Genetics, BMC Genomics, Toxicon and Copeia.
Media Activities
Podcast and article about snakebite for Planet Earth magazine
http://www.nerc.ac.uk/publications/planetearth/2012/spring/planet-earth-spr12.pdf
http://planetearth.nerc.ac.uk/multimedia/story.aspx?id=1101
Research radio interview about venom evolution for the BBC World Service ‘World update: daily commute’ program. Aired 19th September 2012. http://www.bbc.co.uk/podcasts/series/worldupmc
An article about my venom evolution research by award-winning science writer Carl Zimmer for National Geographic
http://phenomena.nationalgeographic.com/2013/01/09/on-the-origin-of-venom
Press articles on my research have been featured by news agencies such as the BBC, The Economist and Fox News
http://www.bbc.co.uk/news/health-19634621
http://www.economist.com/blogs/babbage/2012/10/evolution-venom
http://www.foxnews.com/health/2012/09/20/snake-venom-could-be-used-to-develop-drugs-for-cancer-diabetes
Research
Research Interests
Elucidating the venom composition of venomous animals

Venoms are an important trait in the animal kingdom and have evolved on many occasions in different animal lineages. I am interested in utilising molecular tools, including next-generation sequencing, to elucidate the genes expressed in the venom glands of fish, cnidarians and snakes. Coupling gene expression with proteomic tools allows us to investigate which of these genes are present in the secreted venom of the animal and which cause pathology when the venom is used for predation or defence against natural prey or predators and humans.
Reconstructing the origin of the venom trait in fish

Despite representing the largest group of venomous vertebrates after reptiles, very little is known about fish venom. Fish venoms appear to have evolved independently on many occasions, including multiple times in both bony and cartilaginous fish, and are used for protection the animal against predation. I am using a multi-disciplinary approach to identify toxin components in the venoms of these independent fish lineages to use them as characters to reconstruct the evolutionary origin of the venom trait in this large vertebrate lineage. This NERC funded project will also permit comparisons of the relative complexity of venoms from defensively and predatory venomous animals.
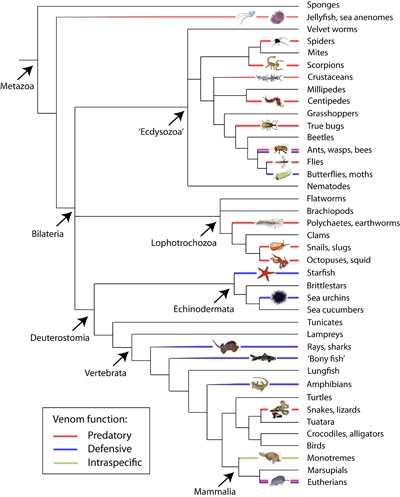
Multi-locus gene family evolution and sites of expression in venomous reptiles
The venoms of reptiles, particularly many snakes, contain numerous protein and peptide components (toxins) that are often encoded by multi-locus gene families. Such gene families are responsible for the production of multiple, related, toxin components thanks to the duplication of genes. I am particularly interested in the mode and tempo of evolution in these gene families, because of their often extreme and rapid nature. Interestingly, venom-related genes appear to have evolved from physiological genes important for homeostasis, and we recently have demonstrated that this process is reciprocal, with some physiological genes evolving from venom toxins. I am interested in exploring the genetic mechanisms responsible for this dynamic phenomenon, using genomic and transcriptomic molecular data.
Development and testing of snake antivenoms
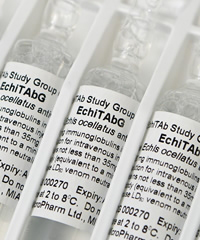
Venomous snakebite is a serious medical concern, particularly in areas of the tropics. Snakebite mortality is thought to approach approximately 100,000 cases worldwide per year, resulting in the World Health Organisation classifying snakebite as a neglected tropical disease. Antivenoms are the only specific treatment for snakebite - they work by containing immunoglobulins that are specific to the toxins found in venom, allowing them to bind and neutralise the activity of the venom. In collaboration with colleagues at the Liverpool School of Tropical Medicine and MicroPharm Ltd I have been investigating methods to improve the safety, stability, efficacy and geographical scope of existing antivenom products, alongside developing new methods to produce ‘next-generation antivenoms’.

Publications
2013
Conlon JM, Attoub S, Arafat H, Mechkarska M, Harrison RA, Casewell NR, Calvete JJ. 2013. Cytotoxic activities of [Ser49] phospholipase A2 from the venom of the saw-scaled vipers Echis ocellatus, E. coloratus, E. pyramidum leakeyi, E. carinatus sochureki. Toxicon. In press.
Casewell NR, Wüster W, Vonk FJ, Harrison RA, Fry BG. 2013. Complex cocktails: the evolutionary novelty of venoms. Trends in Ecology & Evolution 28, 219-229. doi: 10.1016/j.tree.2012.10.020.
Brust A, Sunagar K, Undheim EAB, Vetter I, Yang D, Casewell NR, Ruder T, Jackson TNW, Koludarov I, Alewood PF, Hodgson WC, Lewis RJ, King GF, Antunes A, Hendrikx I, Fry BG. 2013. Differential evolution and neofunctionalization of snake venom metalloproteinase domains. Molecular & Cellular Proteomics 12, 651-663. doi: 10.1074/mcp.m112.023135.
Jackson T, Casewell NR, Fry BG. 2013. Response to “Replies to Fry et al. (Toxicon. 2012. 60(4), 434-448). Part B. properties and biological roles of squamate oral products: the “venomous lifestyle and preadaptation”. Toxicon 64, 113-115. doi: 10.1016/j.toxicon.2012.06.001.
Jackson T, Casewell NR, Fry BG. 2013. Response to “Replies to Fry et al. (Toxicon. 2012. 60(4), 434-448). Part A. Analyses of squamate reptile oral glands and their products: a call for caution in formal assignment of terminology designating biological function”. Toxicon 64, 106-112. doi: 10.1016/j.toxicon.2012.11.006.
Al-Abdulla I, Casewell NR, Landon J. 2013. Long-term physicochemical and immunological stability of a liquid formulated intact ovine immunoglobulin-based antivenom. Toxicon 64, 38-42.doi: 10.1016/j.toxicon.2012.12.022.
2012
Casewell NR, Huttley GA, Wüster W. 2012. Dynamic evolution of venom proteins in squamate reptiles. Nature Communications 3, 1066. doi: 10.1038/ncomms2065.
Casewell NR. 2012. On the ancestral recruitment of metalloproteinases into the venom of snakes. Toxicon 60(4), 449-454. doi: 10.1016/j.toxicon.2012.02.006.
Fry BG, Scheib H, Junqueira-de-Azevado ILM, Silva DA, Casewell NR. 2012. Novel transcripts in the maxillary venom glands of advanced snakes. Toxicon 59(7-8), 696-708.
Fry BG, Casewell NR, Wüster W, Vidal N, Young B, Jackson T. 2012. The structural and functional diversification of the Toxicoferan reptile venom system. Toxicon 60(4), 434-448. doi: 10.1016/j.toxicon.2012.02.013.
2011
Casewell NR, Wagstaff SC, Harrison RA, Renjifo C, Wüster W. 2011. Domain loss facilitates accelerated evolution and neofunctionalization of duplicate snake venom metalloproteinase toxin genes. Molecular Biology and Evolution28(9): 2637-2649. doi: 10.1093/molbev/msr091.
Casewell NR, Wagstaff SC, Harrison RA, Wüster W. 2011. Gene tree parsimony of multi-locus snake venom protein families reveals species tree conflict as a result of multiple parallel gene loss. Molecular Biology and Evolution 28(3): 91-110. doi: 10/1093/molbev/msq302.
Harrison RA, Cook DA, Renjifo C, Casewell NR, Currier RB, Wagstaff SC. 2011. Research strategies to improve snakebite treatment: challenges and progress. Journal of Proteomics 74(9): 1768-1780. doi: 10.1016/j.jprot.2011.06.019.
Williams DJ, Gutiérrez J-M, Calvete JJ, Wüster W, Ratanabanangkoon K, Paiva O, Brown N, Casewell NR, Harrison RA, Rowley P, Jensen SD, Winkel KD, Warrell DA. 2011. Ending the drought: new strategies for improving the flow of affordable, effective antivenoms in Asia and Africa. Journal of Proteomics 74(9): 1735-1767. doi: 10.1016/j.jprot.2011.05.027.
2010
Casewell NR, Cook DAN, Wagstaff SC, Nasidi A, Durfa N, Wüster W, Harrison RA. 2010. Pre-clinical assays predict pan-African Echis viper efficacy for a species-specific antivenom. PLoS Neglected Tropical Diseases 4(10): e851. doi:10.1371/journal.pntd.0000851.
2009
Casewell NR, Harrison RA, Wüster W, Wagstaff SC. 2009. Comparative venom gland transcriptome surveys of the saw-scaled vipers (Viperidae: Echis) reveal substantial intra-family gene diversity and novel venom transcripts. BMC Genomics 10: 564. doi:10.1186/1471-2164-10-564.
