Staff Profile of Dr Richard Challis

- Name
- Dr Richard Challis
- Position
- Postdoctoral Research Officer
- r.challis@bangor.ac.uk
- Phone
- +44(0)1248 383822
- Location
- 3rd Floor, ECW Building, School of Natural Sciences, Bangor University, Deiniol Road, Bangor, Gwynedd, LL57 2UW, UK
Research
Research
Genomics of Speciation and Adaptive Radiation
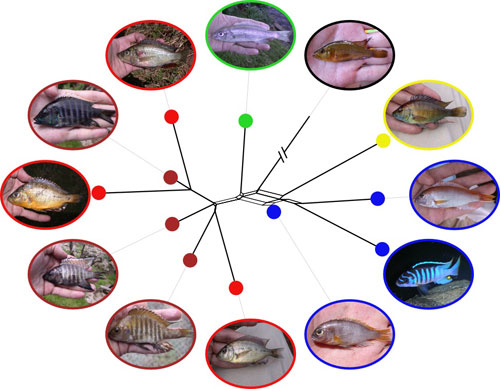
My current research in George Turner's lab is making use of growing genomic resources for cichlid fish together with next-generation sequencing to elucitated mechanisms and processes in speciation and adaptive radiation. At a broad scale, I have been investigating the prevalance of hybridisation between distinct lineages within the large-scale Lake Malawi adaptive radiation using sequenced AFLP markers in collaboration with Martin Genner. On a much smaller scale, our attentions have recently turned to incipient speciation events in volcanic crater lakes for which we now have full genome sequence data through collaboration with Eric Miska, Richard Durbin and their PhD student Milan Malinsky. The genomic resolution that we have achieved between such recently diverged morphs promises to shed light on the behavioural, ecological and genomic processes underlying cichlid speciation and adaptive radiation.
Comparative Genomics of Shoot Branching
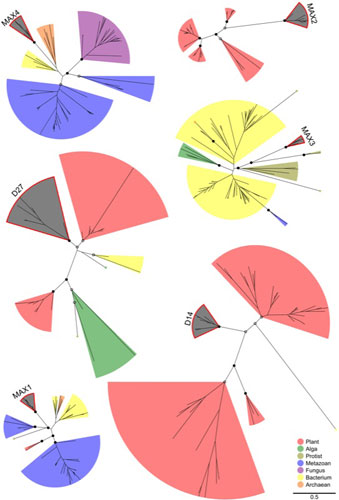
Plants are stationary organisms and respond to environmental factors through developmental plasticity, particularly by controlling shoot-branching through the development, dormancy and outgrowth of buds. I had a BBSRC funded postdoc in Ottoline Leyser's lab using comparative genomics to investigate the hormonal control of shoot branching as part of an ERA-Net Plant Genomics project with collaborators in France, Germany, the Netherlands and Spain. I analysed project data including genomic, microarray and trait association data and investigated semantic approaches to automate bioinformatic analyses. I concentrated on genes in the synthesis and signalling pathway of the hormone stigolactone (SL) and comparative analysis of these genes and their wider gene families revealed patterns of recruitment to the SL pathway.
Evolution of Oak Gallwasp Communities
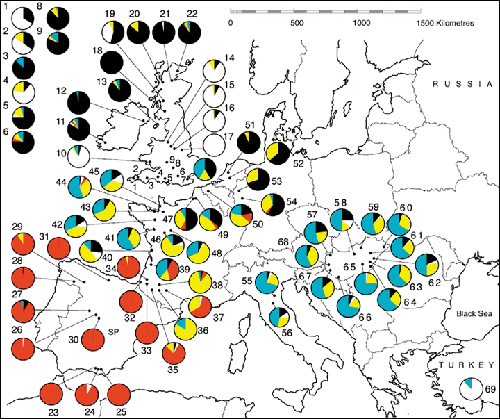
My PhD research, supervised by Graham Stone and made possible by collaborators throughout the Western Palaearctic and beyond, used phylogeography and DNA barcoding to investigate the evolution of oak gallwasp communities. Oak gallwasps have complex lifecycles, alternating between sexually reproducing and parthenogenetic generations that can differ considerably in appearance and host species galled. DNA barcoding is essential to link the two generations, which traditional taxonomy often placed in separate species in order to fully understand gallwasp ecology. Using DNA sequencing I found evidence of cryptic speciation with divergent lineages barely distinguishable morphologically and inducing the formation of highly similar galls. My phylogeographical analyses revealed a dramatic impact of human activity on Andricus kollari in which the entire basal clade had been translocated from its native range in the Eastern Mediterranean to the UK where it's galls were imported in the 1830s as a source of tannins for the manufacture of ink.
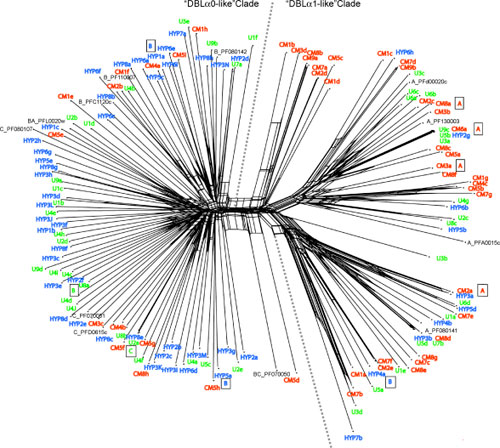
var Gene Expression in Plasmodium falciparum Malaria
This work was part of a study into the link between the pathogenicity of malarial infections and the expression of Plasmodium falciparum variant erythrocyte surface antigens (PfEMP1), led by Alex Rowe. I undertook a phylogenetic analysis of the var gene family that encodes PfEMP1 antigens that supports the division of genes expressed in isolates from individuals with more severe forms of malaria (red) into a separate clade from those taken from individuals with milder symptoms (blue/green). Overall, the study suggests that differences in the cytoadhesive and immuno-modulatory roles of the genes in these two clades is likely to contribute to the differences in pathogenicity of malarial infections.
Evolution of Spider Silk
I undertook a comparative analysis of available spider silk sequence data for my undergraduate dissertation project, supervised by Sara Goodacre and Godfrey Hewitt. We found a high degree of sequence conservation in the C-termini of each major class of silk protein (the highly elastic flagelliform and relatively inelastic spidroin silk proteins). Despite the amino acid divergence, both classes have similar predicted physical properties in this region, suggesting that it is responsible for shared properties of the two classes, perhaps during silk production and protein folding.

